|
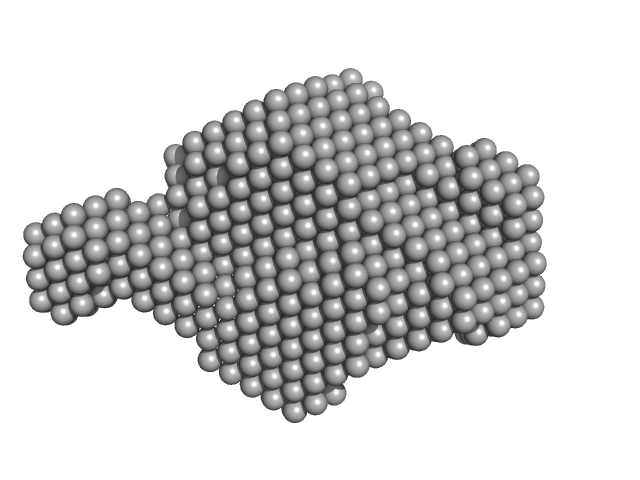
X-ray synchrotron radiation scattering data from solutions of N-terminally truncated Aureochrome 1a in 20 mM HEPES 100 mM NaCl, 10 mM MgCl2, 5% w/v glycerol were collected at the X12SA cSAXS beamline at the Swiss Light Source (Villigen, Switzerland) using a Pilatus 2M detector (s = 4π sin θ/λ, where 2θ is the scattering angle). Samples were filled and mounted in 1 mm quartz capillaries in the dark and kept at 10°C throughout the experiments. Data acquisition using 11.2 keV photons was performed in 500 µm steps along the capillary with 10 × 0.5 s exposure at 20 different positions. SAXS data was collected in the dark from the buffer alone and from the protein using identical positions along the same capillary. For data analysis, all diffraction images were azimuthally integrated, averaged and the buffer signal was subtracted from that of the protein solution to produce the scattering profile displayed in this entry. Molecular weight estimates of N-terminally truncated Aureochrome 1a were derived from the particle volume using Porod or ab initio (DAMMIN) volume estimates. The model displayed above is a refined bead model determined from the spatial alignment of 10 individual DAMMIN shape reconstructions (average NSD = 0.486) that are available in the full zip archive for this entry.
Concentration = UNKNOWN
|
|
 s, nm-1
s, nm-1