|
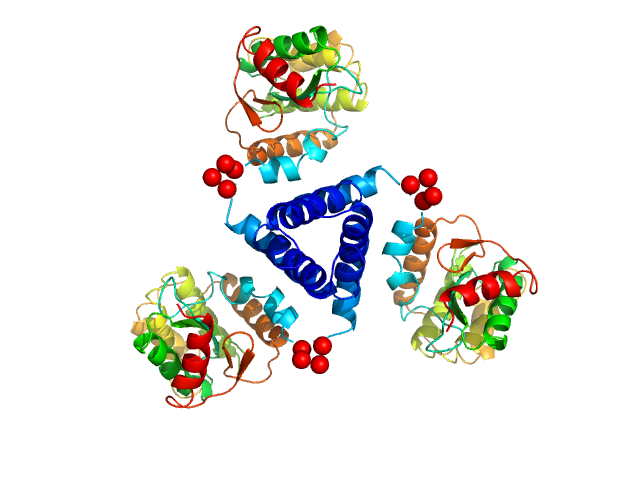
X-ray synchrotron radiation scattering data from solutions of Suppressor of Copper Sensitivity C protein (ScsC) from Proteus mirabilis in 25 mM HEPES 150mM NaCl, 1mM DTT, pH 7.5 were collected on the SAXS/WAXS beam line of the Australian Synchrotron (Melbourne, Australia) using a 2D Photon counting Pilatus 1M-W pixel detector (s = 4π sin θ/λ, where 2θ is the scattering angle). Fourteen successive 2 second frames were collected across solute concentrations of 2.15-8.85 mg/ml. The SAXS data displayed this entry was derived from a 2.15 mg/ml sample. The data were normalized to the intensity of the transmitted beam and radially averaged and the scattering of the solvent-blank was subtracted. The models and corresponding fits include those derived from rigid-body modelling using CORAL (top) and a representative ensemble of six trimeric ScsC structures determined using ensemble optimization method (EOM). The Rg and Dmax distributions derived from EOM are included in the full entry zip archive.
Wavelength = UNKNOWN
|
|
 s, nm-1
s, nm-1
 Rg, nm
Rg, nm