|
|
|
|
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF tetramer, 223 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jul 7
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
412 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF monomer, 25 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
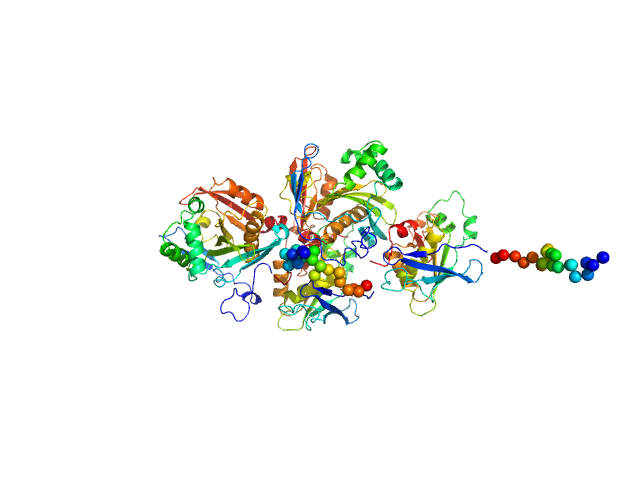
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF (Δ444-462) dimer, 107 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 (KARRIKIN INSENSITIVE 2) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 (KARRIKIN INSENSITIVE 2) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 (KARRIKIN INSENSITIVE 2) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
283 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 (KARRIKIN INSENSITIVE 2) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
4.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
345 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 - S95A (KARRIKIN INSENSITIVE 2 - S95A) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 - S95A (KARRIKIN INSENSITIVE 2 - S95A) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 - S95A (KARRIKIN INSENSITIVE 2 - S95A) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
171 |
nm3 |
|
|