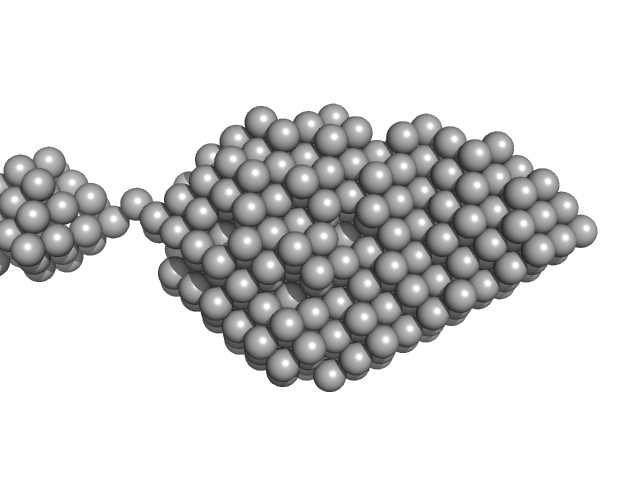
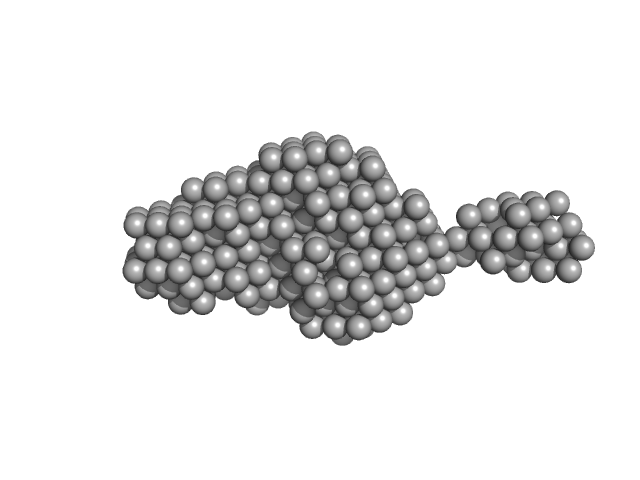
|
Synchrotron SAXS
data from solutions of
Human NEI like DNA glycosylase 1 (NEIL1) bound to Proliferating Cell Nuclear Antigen (PCNA) and DNA
in
25mM HEPES 100mM NaCl 1mM DTT, pH 7.5
were collected
on the
12.3.1 (SIBYLS) beam line
at the Advanced Light Source (ALS) storage ring
(Berkeley, CA, USA)
using a Pilatus3 X 2M detector
at a sample-detector distance of 1.4 m and
at a wavelength of λ = 0.1127 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 0.6 and 2.8 mg/ml were measured
at 10°C.
24 successive
0.200 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
We have attempted to characterize this novel complex using SAXS. This complex shows prominent concentration dependency. Due to presence of DNA as part of this complex, "280nm UV absorbance"-based concentration estimations have been largely compromised, producing un-interpretable forward scattering upon concentration normalization. Thus we have relied upon the "concentration-independent" SAXS-MoW to estimate the MW of the scatterer at physiologically relevant micromolar concentrations. MW estimated using qmax=0.2431 (<=8/Rg) suggests this novel complex to be 86.7kDa, comparable to theoretical value of 85.5 kDa.
|
|
 s, nm-1
s, nm-1