|
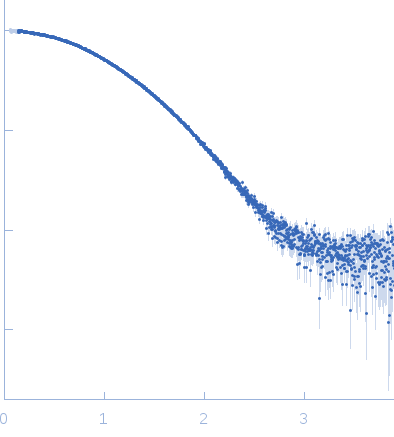
Synchrotron SEC-SAXS data collection was performed at the B21 beamline of the Diamond Light Source (UK) using a Pilatus 2M detector at a sample-detector distance of 4.0 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle). 50 µl purified hTERC 1-20 DNA G-quadruplex at a concentration of 5.58 mg/ml was injected into a 4.6 ml Shodex KW402.5-4F size exclusion column at a flow rate of 0.16 ml/min that was connected to the SAXS flow cell (25°C). The column diluted the sample 3-fold to approximately 2 mg/ml. Each frame was exposed for 3 seconds. 9 frames of the sample peak region were integrated, buffer subtracted and merged using the ScÅtter software package (http://www.bioisis.net/scatter). Guinier Rg, I(0) and their standard uncertainties were determined with GNOM/DATRG from ATSAS suite 2.8.2 (https://www.embl-hamburg.de/biosaxs/software.html). The Porod volume was calculated with DATPOROD from the ATSAS suite. The molecular weight derived from I(0) was calculated with SAXSMoW2 (http://saxs.ifsc.usp.br) and corrected for the appropriate mass density of the G-quadruplex DNA (1.85 g/cm³).
|
|
 s, nm-1
s, nm-1


