|
Synchrotron SAXS data from solutions of HicB bound to HicA in 25 mM Tris 150 mM NaCl, pH 7.5 were collected using in-line SEC-SAXS at the B21 beam line at the Diamond Light Source (Oxfordshire, UK) using a Pilatus 2M detector at a sample-detector distance of 4.0 m and at a wavelength of λ = 0.1 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In line SEC-SAXS was performed using an Agilent 1200 HPLC system equipped with a 2.4 mL Superdex S200 column (GE Healthcare). A sample concentration of 5mg/ml was loaded onto the column at a flow rate of 0.04 mL/min. Data frames were collected at 25°C for the entire column elution with an exposure time of 3 seconds per frame through an angular s-range of 0.04-4 nm-1. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted using data frames measured from the flow-through column buffer with no macromolecular sample present.
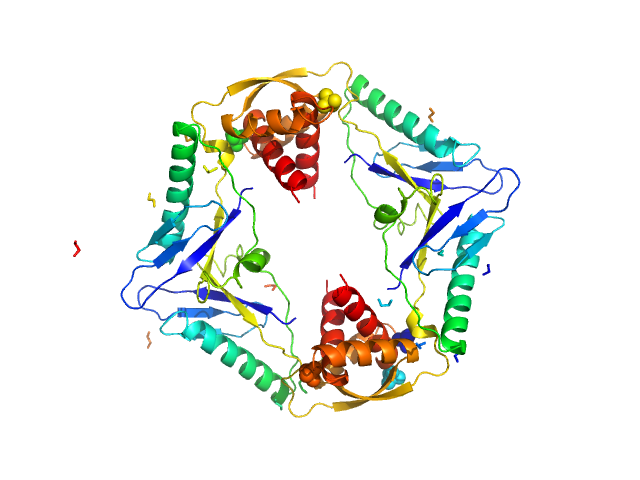
Analysis of the data involved ScÅtter (www.bioisis.net/). The resultant curve was fit with the crystal structure of HicB, PDB Code: 6G26.
|
|
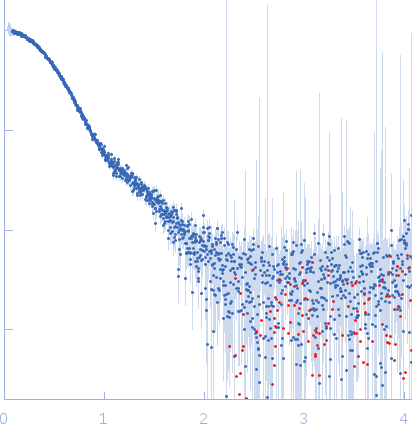 s, nm-1
s, nm-1