|
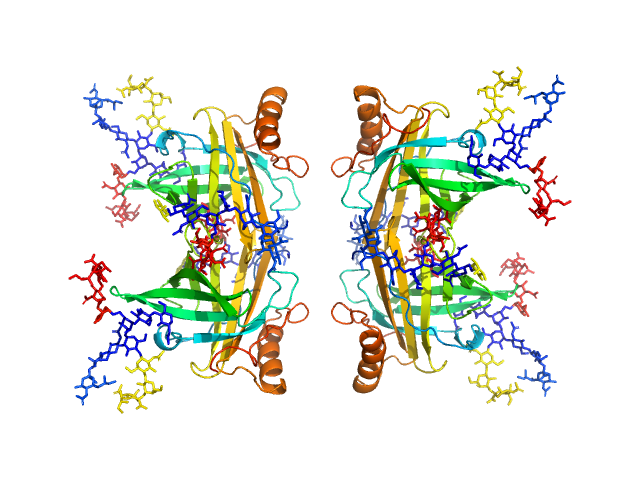
Synchrotron SAXS data from solutions of apolipoprotein D (ApoD) tetramer in 50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH 7.4 were collected on the SAXS/WAXS beam line at the Australian Synchrotron (Melbourne, Australia) using a Pilatus 1M detector at a wavelength of λ = 0.10322 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). The SAXS data were collected using size-exclusion chromatography SAXS (SEC-SAXS).
Additional SEC-SAXS information: SEC-column: GE Healthcare Superdex S200 5/150; Loading concentration (measured using BCA assay): 7.5 mg/ml; Injection volume : 50 µl; Flow rate: 0.45 ml/min; Number of frames averaged over the elution peak: 15. The starting pdb model was the apo-form of apoD with modelled glycans (Oakley et al. 2012). The refined SASREF CV model incorporated contact restraints derived from crosslinking mass spectrometry (crosslinker BS3) that are included in the full entry zip archive (32 Ang between: a, K55-K55; b, K156-K156; c, K155-K156 and; d, K144-K156.)
|
|
 s, nm-1
s, nm-1