|
Synchrotron SAXS data from solutions of the PDZ1-2 domain of postsynaptic density protein 95 (PSD-95) bound to RRESEI peptide in 20 mM TRIS/HCl, 150 mM NaCl + 10 mM RRESEI, pH 8.5 were collected on the EMBL P12 beam line at the PETRA III storage ring (Hamburg, Germany) using a Pilatus 2M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 15.00 mg/ml was measured at 15°C. 15 successive 0.050 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
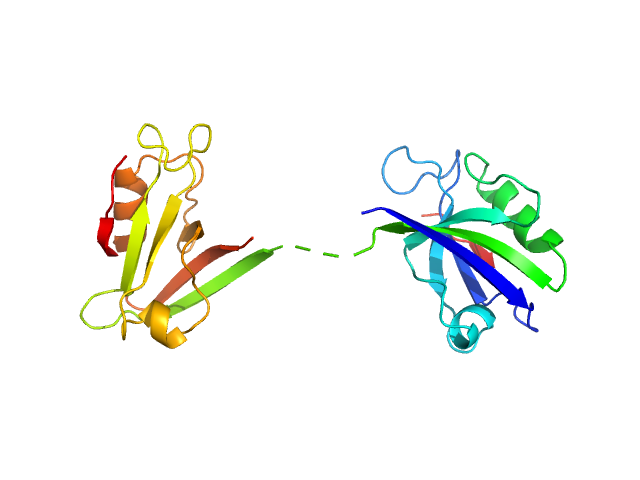
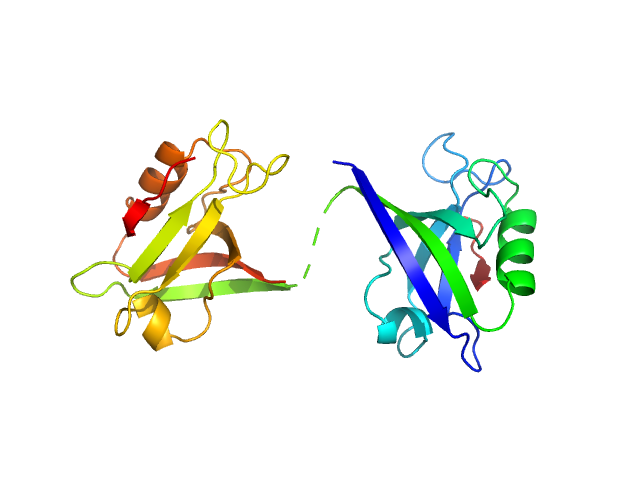
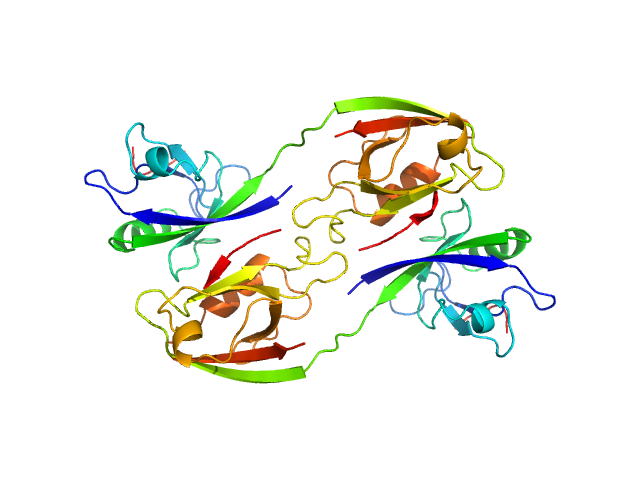
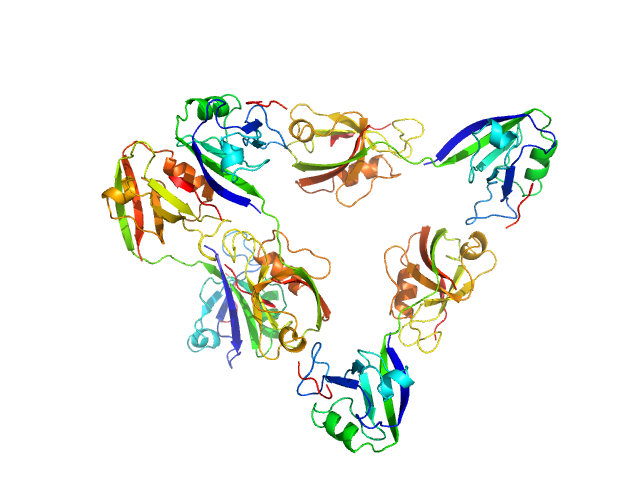
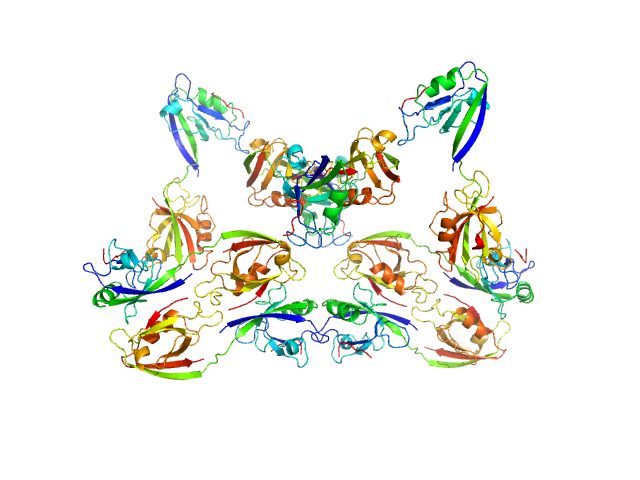
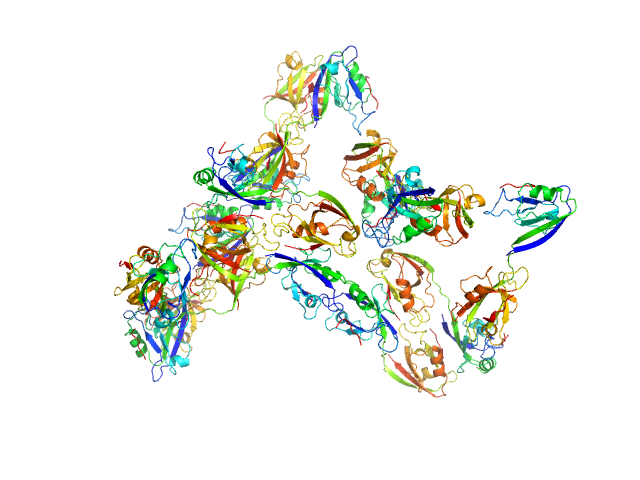
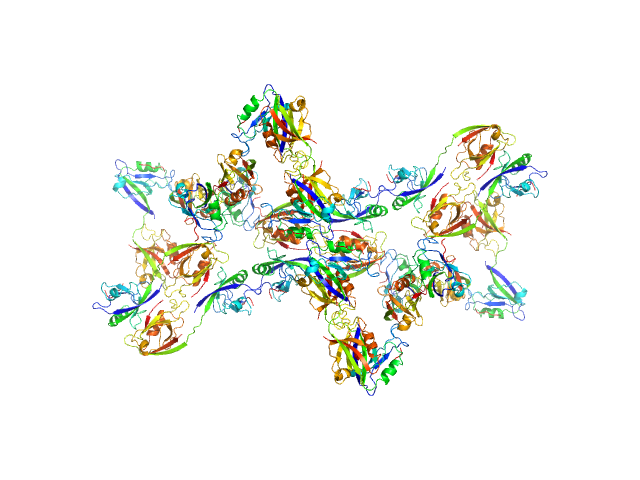
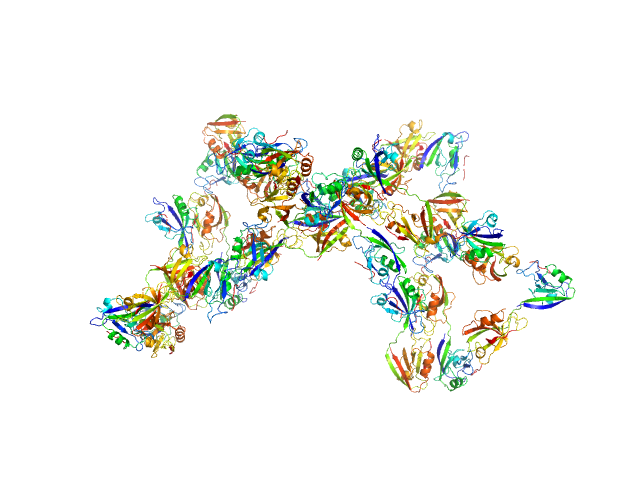
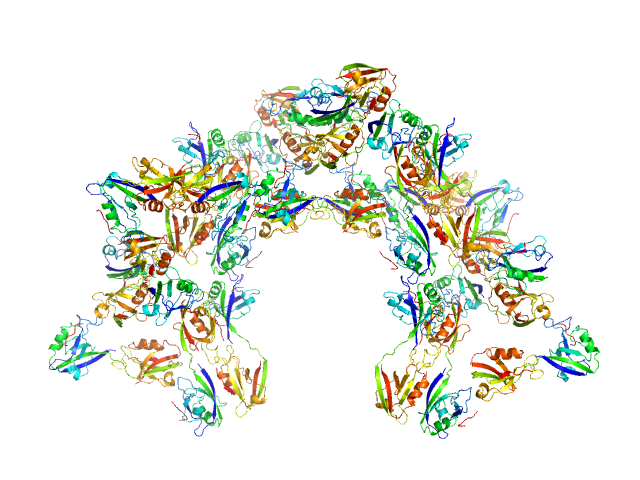
Scattering data are fitted using the ATSAS OLIGOMER program using a suite of models. The ATSAS program FFMAKER was used to generate form factors for each model in the suite. Each model is assigned a volume fraction to fit the observed scattering profile. 3 monomer models are included, the first is a compact conformation of PDZ1-2 similar to PDB entries 6spv/6spz; the other two are domain models obtained from a representative run of the ATSAS EOM program with data projected to infinite dilution. Multimeric models are drawn from a "clustering Spacegroup", unit cell 14.8nm symmetry I2(1)3, and consist of identical copies of an extended conformation of PDZ1-2 (similar to PDB entry 3zrt) assembled by symmetry operations.
In the order of deposition:
Model number; Stoichiometry; MW (kDa); source; Volume fraction.
1; 1; 21; Crystal Structure; 0.112.
2; 1; 21; EOM; 0.186.
3; 1; 21; EOM; 0.656.
4; 2; 42; clustering Spacegroup; 0.000.
5; 4; 84; clustering Spacegroup; 0.000.
6; 8; 168; clustering Spacegroup; 0.034.
7; 12, 252; clustering Spacegroup; 0.005.
8; 16, 336, clustering Spacegroup; 0.000.
9; 20, 420, clustering Spacegroup; 0.000.
10; 24, 504, clustering Spacegroup; 0.006.
|
|
 s, nm-1
s, nm-1