| MWexperimental | 205 | kDa |
| MWexpected | 205 | kDa |
| VPorod | 2150 | nm3 |
|
log I(s)
4.70×10-1
4.70×10-2
4.70×10-3
4.70×10-4
|
 s, nm-1
s, nm-1
|
|
|
|

|
|
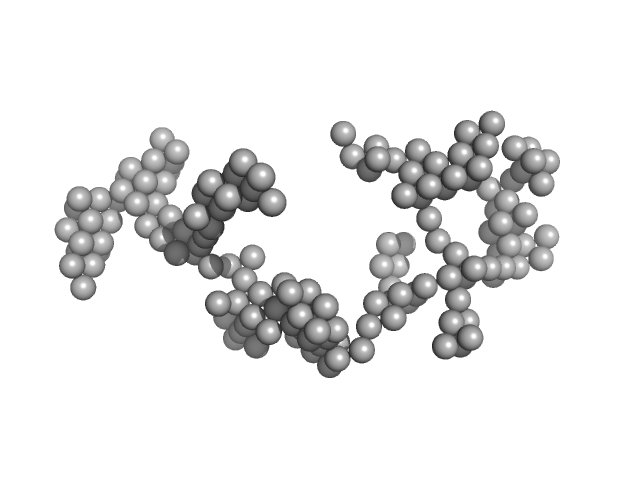
|
|
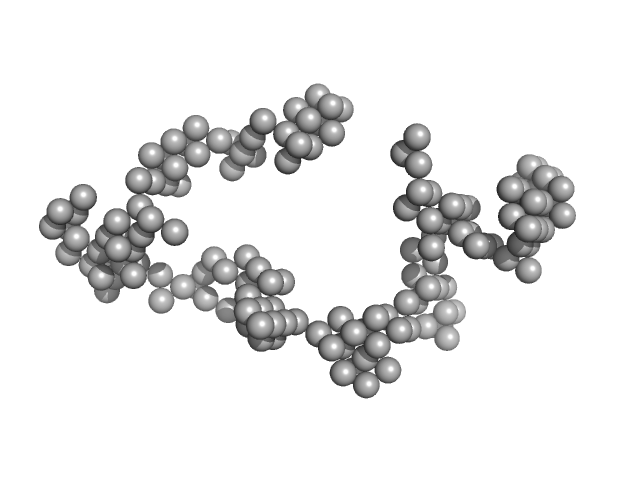
|
|
Synchrotron SAXS
data from solutions of
Braveheart in 0 mM MgCl2
in
50 mM HEPES-KOH, 100 mM KCl, pH 7.6
were collected
on the
B21 beam line
at the Diamond Light Source storage ring
(Didcot, UK)
using a Pilatus 2M detector
at a sample-detector distance of 4 m and
at a wavelength of λ = 0.1 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A sample
at 1.9 mg/ml was injected
onto a column
.
620 successive
3 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The last 46 nucleotides in the sequence are not part of Bvht. These were added as 3' structure cassette for primer extension after SHAPE and DMS probing. SEC-column type and SEC parameters (flow-rate, injection volume) = UNKNOWN; Experimental temperature = UNKNOWN |
|
|||||||||||||||||||||