| MWexperimental | 205 | kDa |
| MWexpected | 205 | kDa |
| VPorod | 2380 | nm3 |
|
log I(s)
2.20×10-1
2.20×10-2
2.20×10-3
2.20×10-4
|
 s, nm-1
s, nm-1
|
|
|
|

|
|
|
|
|
|
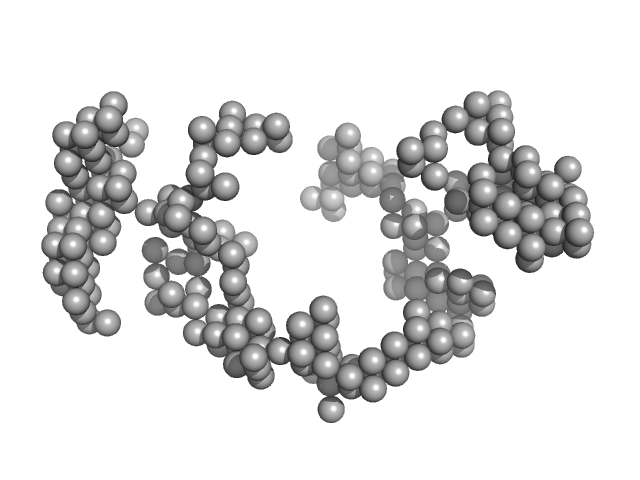
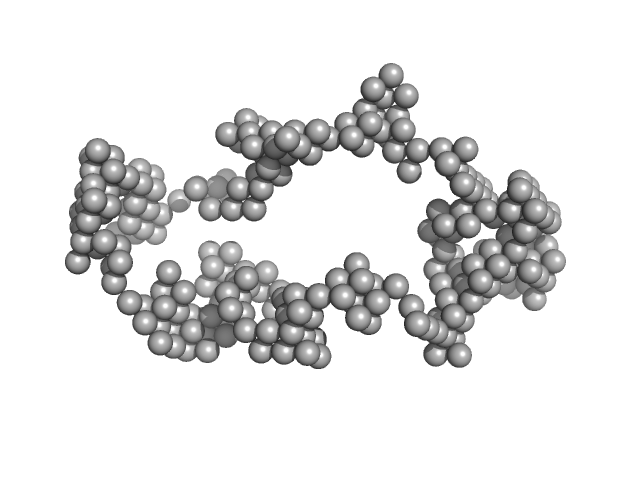
Synchrotron SAXS
data from solutions of
Braveheart in 6 mM MgCl2
in
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH 7.6
were collected
on the
B21 beam line
at the Diamond Light Source storage ring
(Didcot, UK)
using a Pilatus 2M detector
at a sample-detector distance of 4 m and
at a wavelength of λ = 0.1 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A sample
at 2 mg/ml was injected
onto a column
.
616 successive
3 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The last 46 nucleotides are not part of Bvht. These were added as 3' structure cassette for primer extension after SHAPE and DMS probing. SEC-column type and SEC parameters (flow-rate, injection volume) = UNKNOWN; Experimental temperature = UNKNOWN. |
|
|||||||||||||||||||||