|
Synchrotron SAXS data from solutions of the P454 peptide (454-484 residues) of B. Pertussis adenylate cyclase toxin (CyaA) in complex with calmodulin in 20 mM Hepes, 150 mM NaCl, 4 mM CaCl2, pH 7.4 were collected on the SWING beam line at the SOLEIL storage ring (Saint-Aubin, France) using a CCD AVIEX detector at a sample-detector distance of 2.0 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsin θ/λ and 2θ is the scattering angle). 250 successive 1.5 second frames (1second exposure time / 0.5 second dead time) were collected at at 15°C using size-exclusion chromatography SAXS. A 50 μl sample containing 136 µM calmodulin and a 2-fold excess of P454 was injected onto an Agilent Bio SEC-3, 300 Å column and eluted at a 0.20 ml/min flow rate.
Scattered intensities were converted into absolute scale (cm-1) values using the scattering of water. Two independent determinations of the molecular mass were obtained using the programs SAXSMow2 and ScÅtter3 available at the URLs http://saxs.ifsc.usp.br/ and https://bl1231.als.lbl.gov/scatter/, respectively. The average value is MWexperimental=19.5 kDa.
P454-CAM complex: Top panel: Comparison of the experimental data (blue dots) with the calculated scattering pattern (red line) of the DENSS ab initio model shown on the right [Ab initio electron density determination directly from solution scattering data. T. Grant, Nat Methods 15, 191–193 (2018)].
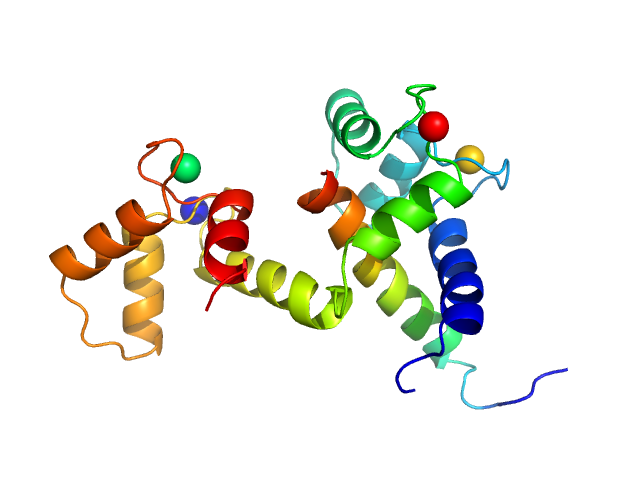
Bottom panel: Comparison of the experimental data (blue dots) with the calculated scattering pattern (red line) of the DADIMODO model shown on the right. chi2=1.49. The program that uses an all atom description of the molecules explores conformational space by random modifications of the internal degrees of freedom phi and psi angles within the calmodulin inter-domain helix. [DADIMODO: a program for refining the structure of multidomain proteins and complexes against small-angle scattering data and NMR-derived restraints. G. Evrard, F. Mareuil, F. Bontems, C. Sizun and J. Perez. J Appl. Crystallogr., 2011, Vol 44(6)]. The program is available at the URL https://dadimodo.synchrotron-soleil.fr.
|
|
 s, nm-1
s, nm-1