|
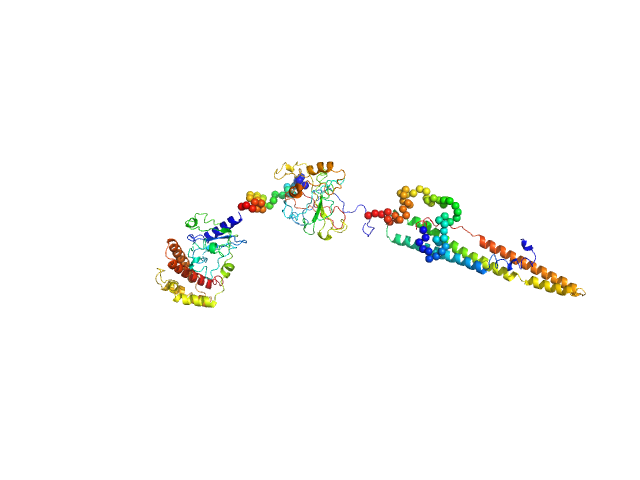
The structural characterisation of the EspK full length protein was performed by SAXS coupled to an online size exclusion chromatography equilibrated with 20 mM Tris-HCl pH 8.0, 300 mM NaCl. The SAXS were collected in the bioSAXS beamline P12-EMBL at DESY Light Source, Hamburg, Germany. Sample consisted of 50 µL at a concentration of 3.6 mg mL-1 and was injected onto a Superdex 200 Increase 3.2/300 size exclusion column attached to a FPLC–Malvern TDA system at a flow rate of 0.1 mL min-1. The elution output was directed through a quartz capillary cell (50 µm thick wall and a 1.7 mm path length) held in vacuum. Data acquisition consisted of 1850 frames (with 1 s exposure time) using a PILATUS 2M detector at the distance of 3.0 m from the sample. Collected two-dimensional images were corrected for variations in beam current, normalized for time exposure, and processed into one-dimensional scattering curves using integrated software at the beamline. Background was manually subtracted using the program Chromix (ATSAS). The first atomic model as obtained by fitting, by ChimeRA, the three part of the protein , obtained by ITASSER, into low resolution SAXS DAMMIF model. This initial model was next refined by Coral, adding the linkers between the middle part with C and N-terminal
|
|
 s, nm-1
s, nm-1