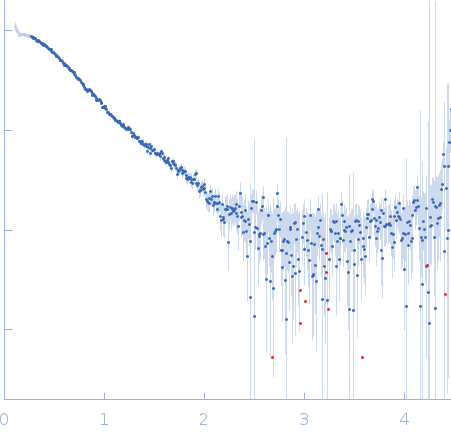
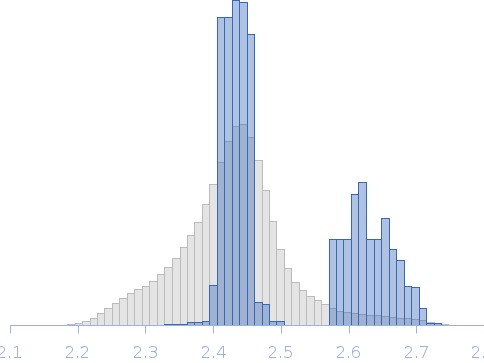
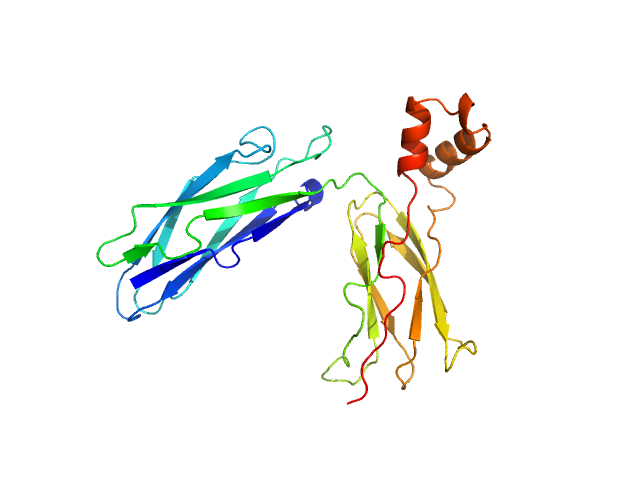
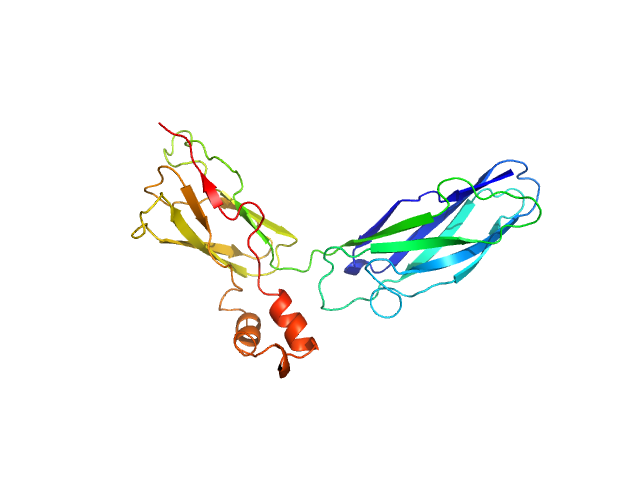
| MWI(0) | 33 | kDa |
| MWexpected | 30 | kDa |
| VPorod | 41 | nm3 |
|
log I(s)
1.16×10-1
1.16×10-2
1.16×10-3
1.16×10-4
|
 s, nm-1
s, nm-1
|
|
|
|
 Rg, nm
Rg, nm
|
|
|
|
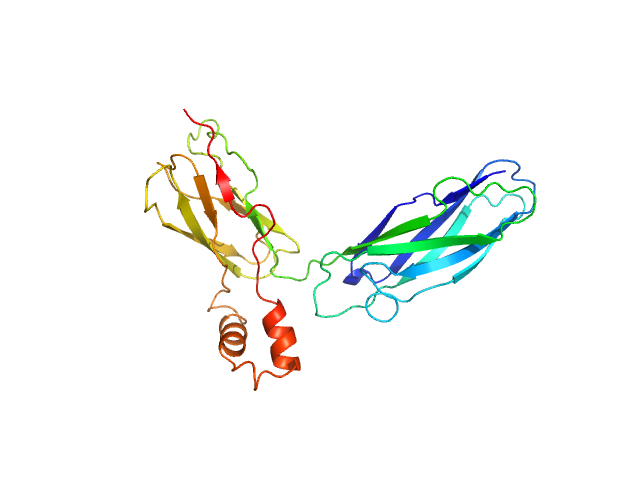
SAXS data from solutions of CBD12 of CALX1.1 in 20 mM Tris, 5 mM β-mercaptoethanol, 200 mM NaCl, 1% v/v glycerol, pH 7.4 were collected using a Xenocs-Xeuss instrument (Institute of Physics, University of São Paulo, Brazil) equipped with a Pilatus detector at a sample-detector distance of 0.8 m and at a wavelength of λ = 0.15418 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 5.30 mg/ml was measured at 24°C. Six successive 1800 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
|
|
|||||||||||||||||||||||||||