|
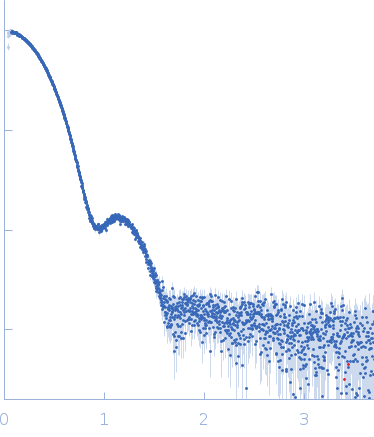
Synchrotron SAXS data from PNPase:2-GMP complex in 20 mM Tris·HCl at pH 8.0, 10 mM NAH2PO4, 60 mM KCl, 1mM MgCl2 and 2mM DTT was collected on the B21-DLS beam-line at the Diamond Light Source (Didcot, UK) using a Pilatus 2M detector at a sample-detector distance of 3.988 m and at a wavelength of λ = 0.1024 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
In-line size-exclusion chromatography (SEC) SAS was employed using an Agilent 1200 HPLC System by injecting a 60 μl sample at 6.4 mg/ml of protein with 417μM of ligand under a flow rate of 0.075 ml/min onto Superdex 200 Increase 3.2/300 column (GE Healthcare) at 25°C collecting 915 successive three-second frames.
The scattering intensities from PNPase:2-GMP complex elution peak region (46-frames) were integrated, buffer subtracted and averaged using the ScÅtter software (version 3Q) to produce the averaged SEC-SAXS profile. From this profile, the pair-wise distance distribution function, P(r), was obtained by indirect Fourier Transform with GNOM (version 5.0) using a momentum transfer range of 0.08990 < s <2.67956 nm-1. The Rg values were estimated by applying the Guinier approximation in the range s < 1.3/Rg.
The displayed SAXS-generated ab initio molecular reconstruction of PNPase:2-GMP complex was obtained using DAMMIF (version 2.3, from ATSAS) - DAMFILT occupancy and volume-corrected bead model - by clustering and averaging dummy residue models from 20 independent runs, with an averaged Normalized Spatial Discrepancy (NSD) of 0.960.
|
|
 s, nm-1
s, nm-1
