|
SAXS data from solutions of PfMDH L-lactate dehydrogenase bound to inhibitor 6a in 100 mM Na-phosphate buffer, 400 mM NaCl, pH 7.4 were measured using a Xenocs Xeuss 2.0 instrument equipped with a Pilatus3 R 1M detector (Department of Macromolecular Physics, Adam Mickiewicz University, Poznań, Poland) at a wavelength of λ = 0.134 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 2.49 mg/ml was measured. Two successive 600 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
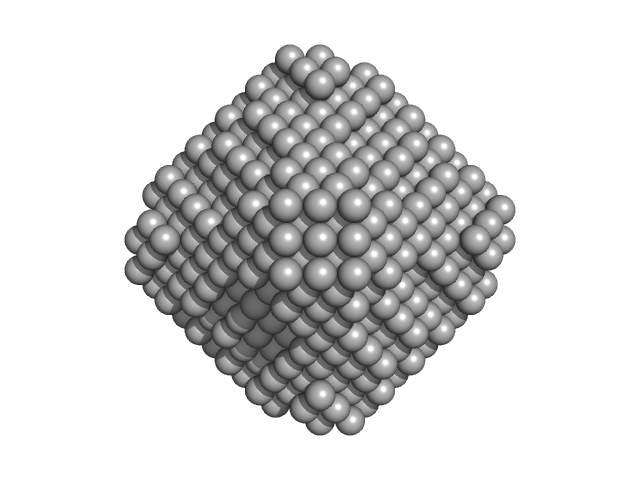
Inhibitor 6a was added to 1 mM concentration. The model displayed is a bead-occupancy and volume-corrected spatial representation derived from an aligned individual model cohort (damfilt) and does not correspond to the fit to the SAXS data displayed in this entry. Experimental temperature: UNKNOWN.
|
|
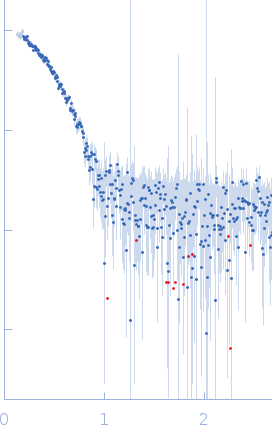 s, nm-1
s, nm-1