|
Synchrotron SAXS data from solutions of the NADase/SLO complex in phosphate buffered saline, pH 7.4 were collected on the 12.3.1 (SIBYLS) beam line at the Advanced Light Source (ALS; Berkeley, CA, USA) using a Pilatus3 X 2M detector at a sample-detector distance of 1.5 m and at a wavelength of λ = 0.103 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 50.00 μl sample at 12 mg/ml was injected at a 0.50 ml/min flow rate onto a Shodex KW-800 series column at 20°C. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted from those sample frames encompassing the SEC elution peak.
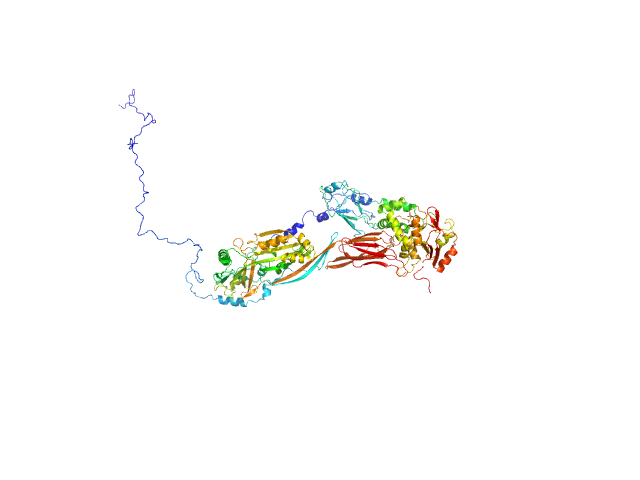
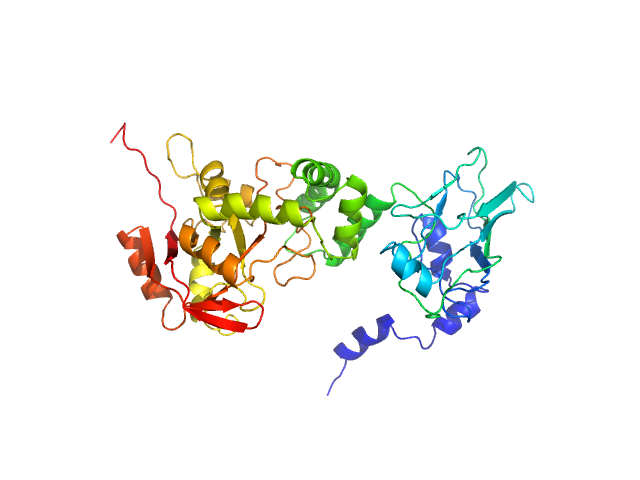
The best fit to the SAXS data is a multistate model consisting of a 9% volume fraction of the compact conformation of the complex (top) and a 53% volume fraction of the extended state (middle), in combination with a 38% volume fraction of free NADase (bottom). Small angle neutron scattering (SANS) with contrast variation data and the subsequent analysis of the contrast variation datasets are made available in the full entry zip archive.
|
|
 s, nm-1
s, nm-1