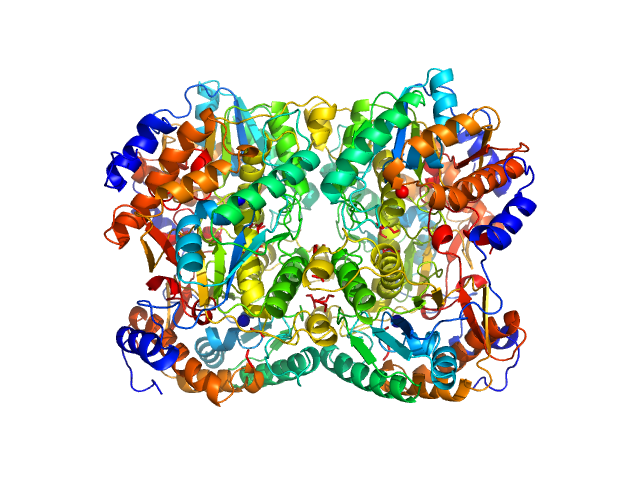
|
Synchrotron SAXS data from solutions of diacetylchitobiose deacetylase in 20 mM TRIS, 200 mM NaCl, pH 7.4 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 30.00 μl sample at 4.7 mg/ml was injected at a 0.35 ml/min flow rate onto a GE Superdex 200 Increase 5/150 column at 20°C. 2880 successive 0.250 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
|
|
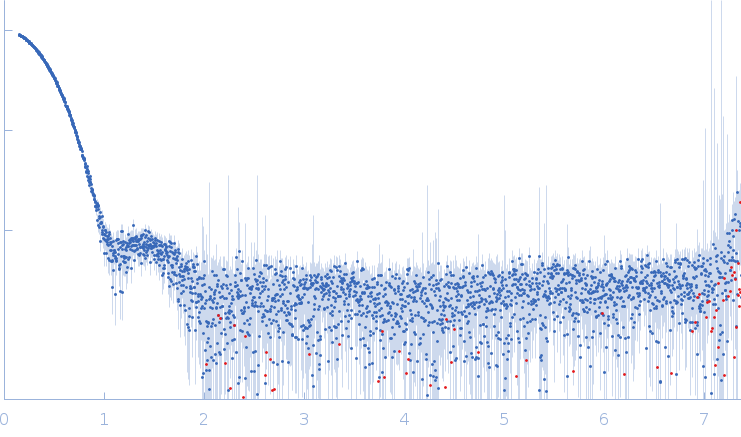 s, nm-1
s, nm-1