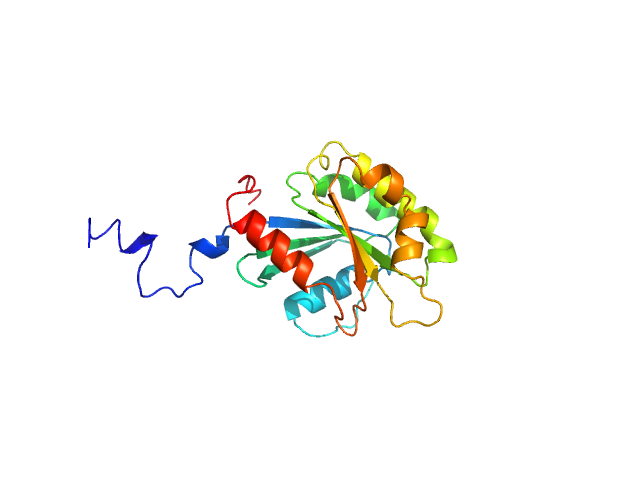
|
GIFT based desmeared and regularized SAXS profile from ADP-ribosylation factor-like protein 15 with GTP in 20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH 8 were collected using an Anton Paar SAXSpace at the CSIR Institute of Microbial Technology (IMTech; Chandigarh, India) equipped with a Mythen 1K detector at a sample-detector distance of 0.3 m and at a wavelength of λ = 0.15414 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 3.00 mg/ml was measured at 10°C. Three successive 1800 second frames were collected. The steps followed to obtain the desmeared and regularized SAXS profile attached were: 1) Line collimation was used for data collection. 2) The smeared raw data was collected for the sample and matched buffer for same exposure time and number of frames. 3) The smeared buffer scattering data was subtracted from that of the smeared sample SAXS data. 4) The program GIFT was used to regularise and desmear the data to obtain the smoothed I(q) profile, corrected for the geometry of the beam profile (representing the scattering modelled as a point collimation source, i.e., the beam profile was used to desmear the original sample-minus-buffer data. 5) The final data profile was scaled to the intensity of the raw sample-minus-buffer intensity profiles and the experimental errors were read-into the smoothed desmeared data file.
|
|
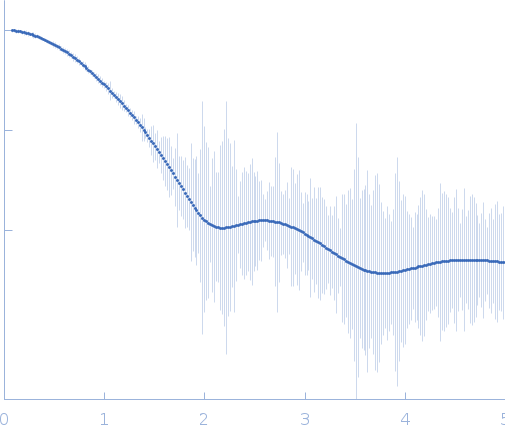 s, nm-1
s, nm-1