|
Synchrotron SAXS data from solutions of calcium-bound calmodulin in 20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH 7.4 were collected on the SWING beam line at the SOLEIL storage ring (Saint-Aubin, France) using a EIGER 4M detector at a sample-detector distance of 2.0 m and at a wavelength of λ = 0.1033 nm (I(s) vs s, where s = 4πsin θ/λ and 2θ is the scattering angle). 1320 successive 1 second frames (0.990 second exposure time / 0.010 second dead time) were collected at 15°C using size-exclusion chromatography SAXS. A 50 μl sample containing 500 µM calmodulin was injected onto a TSKgel G3000SW column (Tosoh Bioscience) and eluted at a 0.50 ml/min flow rate. Scattered intensities were converted into absolute scale (cm-1) values using the scattering of water. Molecular mass was estimated at 15.6kDa using the correlation volume Vc (Rambo, R.P. and Tainer, J. A. (2013) Nature 496(7446):477-81.)
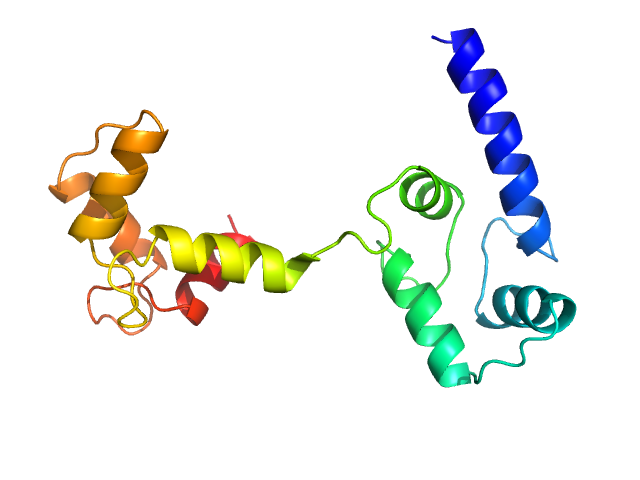
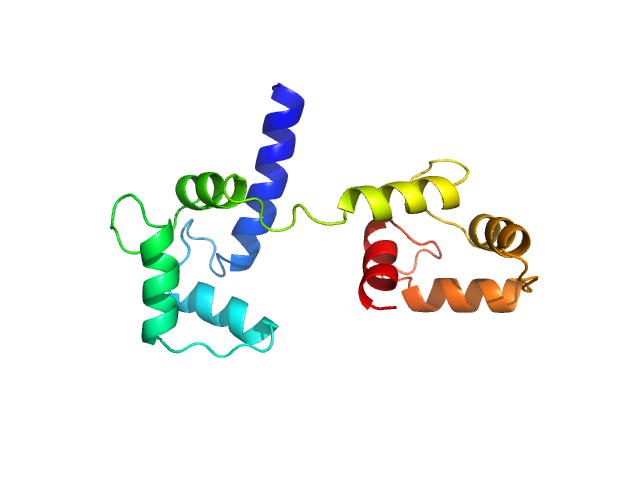
Ab initio model and modelling using an ensemble of conformations:
Bottom panel: Comparison of the experimental data (blue dots) with the calculated scattering pattern (red line) of the DENSS density distribution shown on the right. (Grant, T. D. (2018) Nat Methods 15, 191–193).
Top panel: Comparison of the experimental data (blue dots) with the calculated scattering pattern (red line) of the best EOM ensemble models shown on the right. Chi2 = 1.06. The program Ranch within EOM creates a large (10,000) pool of random conformations, starting from the structures of the two calcium-bound lobes of holo-CaM (PDB ID: 1CLL). Amino acids (76 – 81) in the interlobe linker region were described as dummy residues. We used the native option to generate the linker separating the two lobes of CaM. Dummy residues are subsequently substituted by complete residues using the programs PD2 (Moore, B. L., Kelley, L. A., Barber, J., Murray, J. W., and MacDonald, J. T. (2013) J Comput Chem 34, 1881–9) and SCWRL4 (Krivov, G. G., Shapovalov, M. V., and Dunbrack, R. L. (2009) Proteins 77, 778–95.) before calculating all scattering patterns using CRYSOL. The routine Gajoe within EOM tries to fit the experimental scattering curve by the average of the calculated scattering patterns of an ensemble of conformations using a genetic algorithm protocol. (Tria, G., Mertens, H. D. T., Kachala, M., and Svergun, D. I. (2015) IUCrJ 2, 207-217.)
|
|
 s, nm-1
s, nm-1
 Rg, nm
Rg, nm