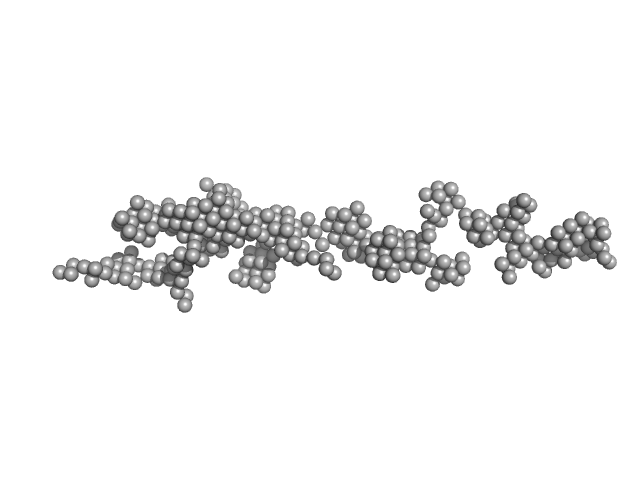
|
Synchrotron SAXS
data from solutions of
Amyloid Beta 1-42 (cluster 1, Set 2, final state)
in
1 mM Hepes, 0.12 % NH4OH, pH 10.7
were collected
on the
EMBL P12 beam line
at the PETRA III storage ring
(DESY; Hamburg, Germany)
using a Pilatus 6M detector
at a sample-detector distance of 3.1 m and
at a wavelength of λ = 0.124 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 2.00 mg/ml was measured
at 20°C.
20 successive
0.195 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
SAXS profile captured during the fibril formation process. The curve belongs to Set 1, cluster 1 and is employed to fit structural kinetics of the process studied with TR-SAXS. This curve was used to obtain and characterise the structural kinetics of cluster 1 studying the process of fibril formation with TR-SAXS. The quoted 'expected MW' is that calculated from the amino acid sequence of the monomeric form of the protein.
|
|
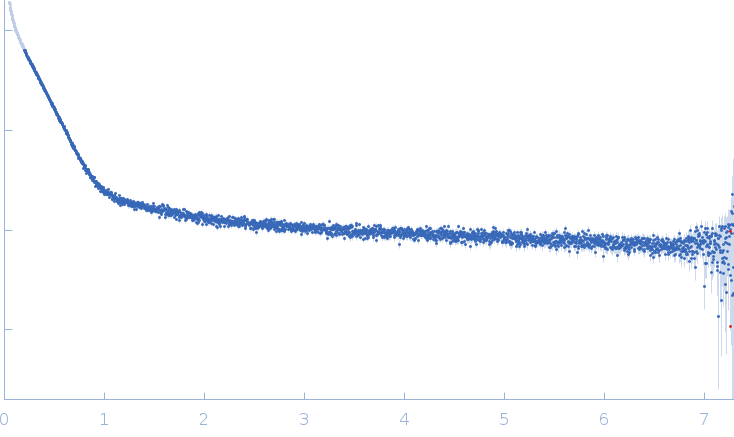 s, nm-1
s, nm-1