| MWexperimental | 72 | kDa |
| MWexpected | 77 | kDa |
| VPorod | 175 | nm3 |
|
log I(s)
1.18×10-1
1.18×10-2
1.18×10-3
1.18×10-4
|
 s, nm-1
s, nm-1
|
|
|
|
|
|
|
|
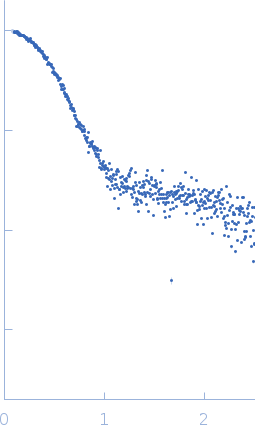
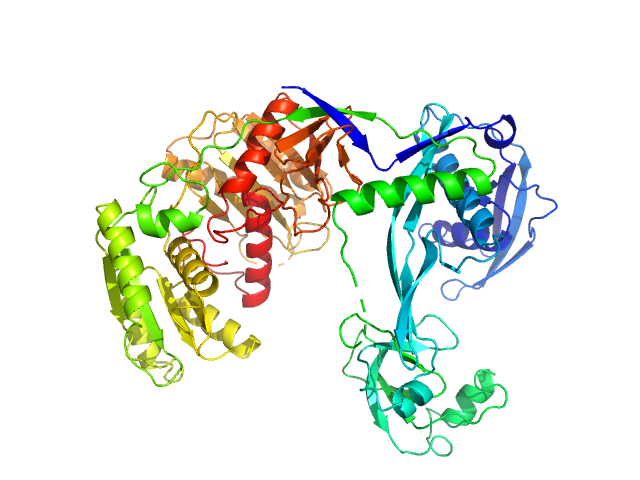
Synchrotron SAXS data from solutions of apo-Thermus thermophilus argonaute protein in 20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH 8 were collected on the BL19U2 beam line at the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) using a Pilatus 1M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.103 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 0.30 mg/ml was measured at 75°C. 20 successive 0.500 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
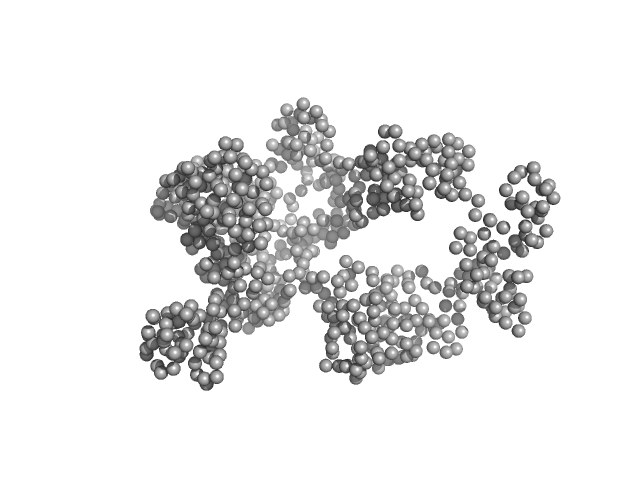
Substitute errors have been used for the ab initio GASBOR modelling (+/-1.0 on each data point). |
|
|||||||||||||||||||||||||||