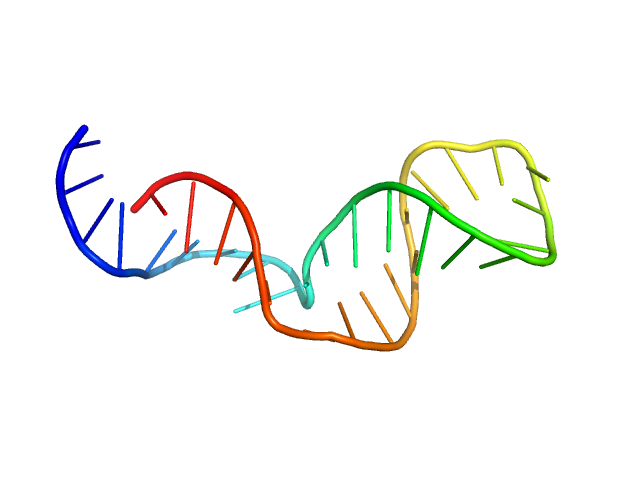
|
Synchrotron SAXS data from Gli-233 were collected at the BM29 BioSAXS beamline at ESRF (France) by the detector Dectris Pilatus 1M. The sample-to-detector distance was 2.849 m, the wavelength λ of the X-Ray beam was 0.09919 nm. The aptamer was diluted to the concentrations of 0.8, 4.0, and 8.2 mg ml-1 in the DPBS buffer with calcium and magnesium. Additional SAXS measurements in the solution without Ca2+ and Mg2+ ions (concentrations of Gli-233 were 0.8 and 4.0 mg ml-1) were carried out to check the influence of their presence on the structure formation of the aptamer. Before SAXS, all aptamer solutions were heated to 90°C and then cooled to 4°C. The recorded range of momentum transfer was 0.1 < s < 3.5 nm-1 (is defined as s = 4π sin(θ) / λ, where 2θ is the scattering angle). Data from the three SAXS curves were extrapolated to zero concentration to exclude the concentration-dependent influence on the data.
|
|
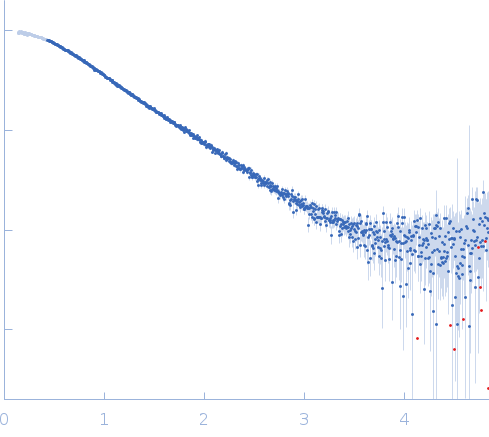 s, nm-1
s, nm-1