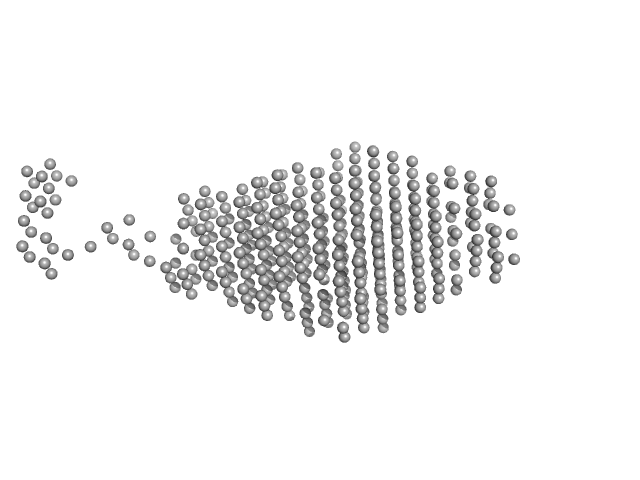
|
Synchrotron SAXS
data from solutions of
Dark-state LOV-activated diguanylate cyclase
in
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH 8
were collected
on the
BM29 beam line
at the ESRF storage ring
(Grenoble, France)
using a Pilatus3 2M detector
at a sample-detector distance of 2.9 m and
at a wavelength of λ = 0.125 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 1.30 mg/ml was measured
at 20°C.
10 successive
1 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Sample preparation and measurment performed under non-actinic conditions (orange/red light only). Sample measurement in batch mode.
|
|
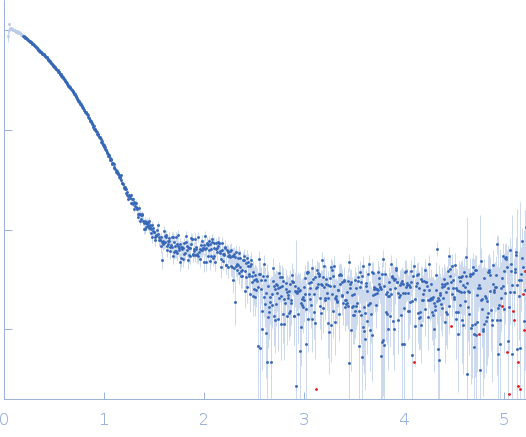 s, nm-1
s, nm-1