|
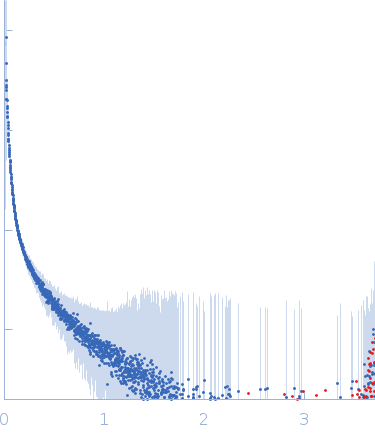
Synchrotron SAXS data from solutions of single self-amplifiyng RNA in 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH 6.1 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 6 m and at a wavelength of λ = 0.123985 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 0.20 mg/ml was measured at 20.2°C. 20 successive 0.100 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
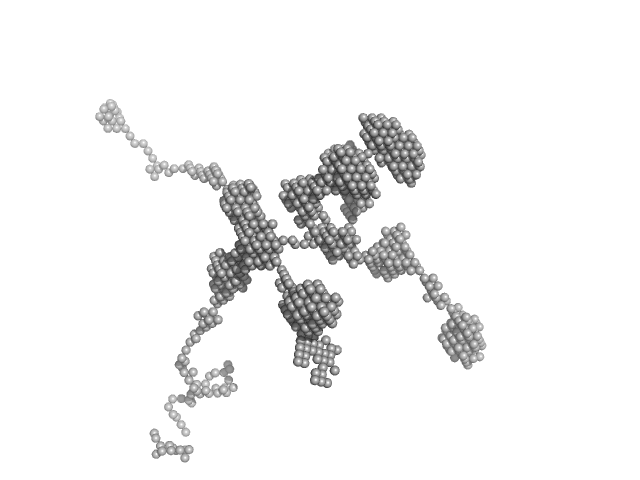
The smin is too large to have a reliable Guinier approximation. Instead, values are approximations from the accessible angular range. Using an alternative approach, modelling with a worm-like chain form factor, a Kuhn length of 12 nm and a contour length of 2270 nm were obtained. DAM volume: 57.4 * 10³ nm³. SAXS and additional AF4 (asymmetrical-flow field flow fractionation) measurements reveal that saRNA is present in a range of different extended molecular conformations. Consequently, the ab initio model represents an effective excluded volume rather than a defined state.
|
|
 s, nm-1
s, nm-1