|
Synchrotron SAXS data from solutions of 22mer poly-ADP ribose (PAR22) in 20 mM Tris-HCl, 1 mM MgCl2, 100 mM NaCl, pH 7.4 were collected on the 16-ID (LiX) beam line at the National Synchrotron Light Source II (NSLS-II; Upton, NY, USA) using a Pilatus3 S 1M detector at a sample-detector distance of 3.7 m and at a wavelength of λ = 0.08188 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 0.5 and 0.7 mg/ml were measured at 25°C. 10 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The low angle data collected at lower concentrations were extrapolated to infinite dilution and merged with the higher concentration data to yield the final composite scattering curve.
ADP ribose: https://pubchem.ncbi.nlm.nih.gov/compound/ADP-Ribose.
Models were generated using all atom molecular dynamics, then running GAJOE to identify a conformational ensemble that fits the data, then performing spectral clustering to identify 4-5 conformations that are representative of the entire conformation. Models are coarse grained such that every point is the mean position of every pair of phosphorus atoms along the backbone. Variances and prevalence across the ensembles are also shown. The corresponding .txt files with xyz coordinates of all the coarse grained models for the entry are made available in the downloadable zip archive.
|
|
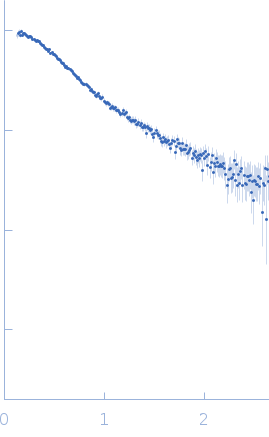 s, nm-1
s, nm-1
![Static model image 22mer of adenosine diphosphate ribose (poly-ADP ribose) OTHER [STATIC IMAGE] model](/media//pdb_file/SASDSM5_fit1_model1.png)