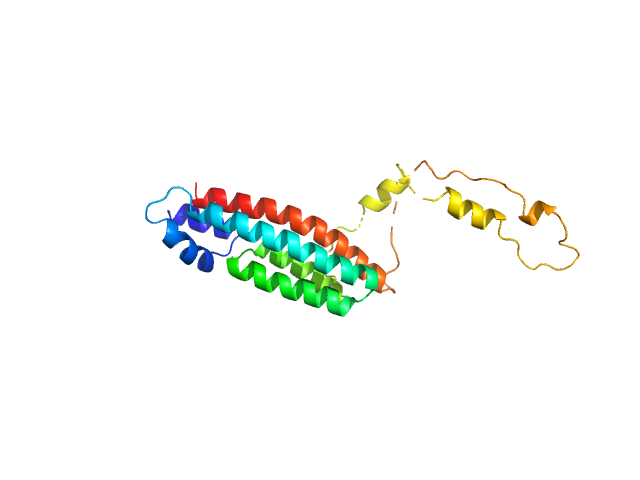
|
Synchrotron SAXS data from solutions of Histidine-phosphotransferase in 50 mM Tris-HCl pH 8, 300 mM NaCl, were collected on the BM29 beam line at the ESRF (Grenoble, France) using a Pilatus3 2M detector at a sample-detector distance of 2.8 m and at a wavelength of λ = 0.099 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 50.00 μl sample at 8 mg/ml was injected at a 0.07 ml/min flow rate onto a GE Superdex 75 Increase 3.2/300 column at 20°C. 1438 successive 2 second frames were collected across the entire SEC elution, where 76 sample frames were selected through the main SEC peak and used for averaging. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Poorly defined Guinier region.
|
|
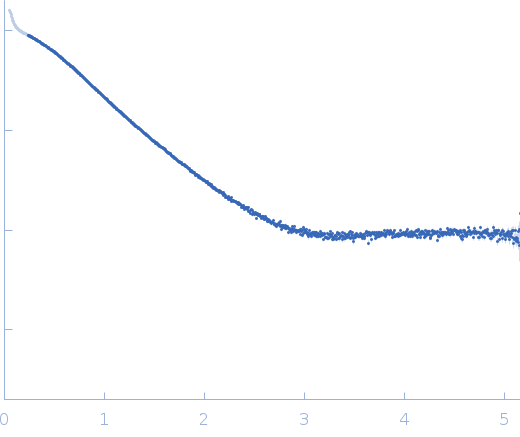 s, nm-1
s, nm-1