|
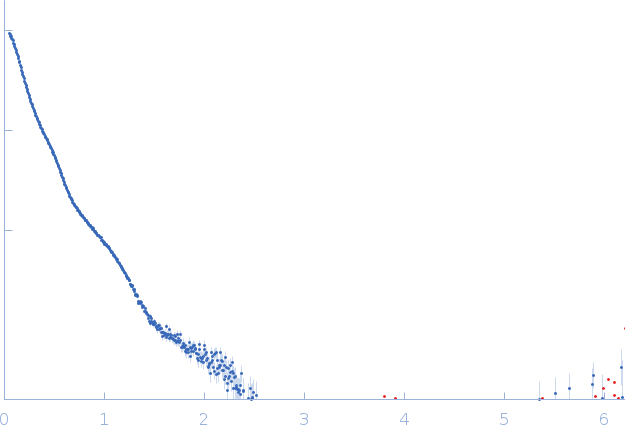
SAXS data from solution of Mouse Fc fragment of IgG binding protein (Fcgbp) in 25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH 7.4 were collected on the BM29 beamline at the ESRF (Grenoble, France) using a Pilatus3 2M detector at a sample-detector distance of 2.8 m and at a wavelength of λ = 0.0826 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Scattering data was collected at 20°C in batch mode and two concentrations (0.12 - 1mg/mL) were merged to generate the presented data. Ten frames of each sample were collected with an exposure time of 1 second per frame. Data reduction and 1D scattering intensities of the sample were performed as per standard BM29 beamline protocols. Ten data frames for each concentration were processed (averaged and buffer subtracted) using primusqt from the ATSAS 3.0.0 package. The radius of gyration (Rg), forward scattering (I(0)), maximum particle dimension (Dmax) and the distance distribution function were determined using Primusqt and GNOM (v5).
This molecule is a secreted protein (produced in CHO-K1) which presents glycosylations (9-11 predicted N-glycosylation sites per monomer). The standard N-glycosylations for CHO-K1 cells are 3.8 kDa per site and around 1 kDa for O-glycosylation. The presence of glycansylations in this molecule can explain the increase of molecular weight calculated from the scattering data using the Bayesian inference approach (679.1-829.45 kDa).
The single ab initio model was obtained by dummy atom modeling using DAMMIN.
|
|
 s, nm-1
s, nm-1
