|
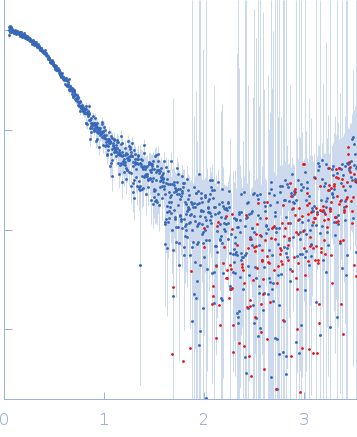
Synchrotron SAXS data from solutions of Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein L177E mutant in 20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT were collected on the BioCAT 18ID beam line at the Advanced Photon Source (APS), Argonne National Laboratory (Lemont, IL, USA) using a Pilatus3 X 1M detector at a sample-detector distance of 3.6 m and at a wavelength of λ = 0.1033 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS with co-flow was employed. The SEC parameters were as follows: A 200.00 μl sample at 4.5 mg/ml was injected at a 0.60 ml/min flow rate onto a Cytiva Superdex 200 Increase 10/300 column at 22°C. 1208 successive 0.500 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
P-Rex1 is a nucleotide exchange factor for Rac GTPases and is mainly expressed in neutrophils where it plays a critical role in cell migration by activation of GTPases. P-Rex1 is reported to be over-expressed in different cancers and therefore it is a potential anti-cancer target, making it important to understand its working mechanism. In unstimulated conditions, P-Rex1 remains auto-inhibited owing to intra-domain interactions and its activation is dependent upon binding to PIP3 and Gbg subunits.
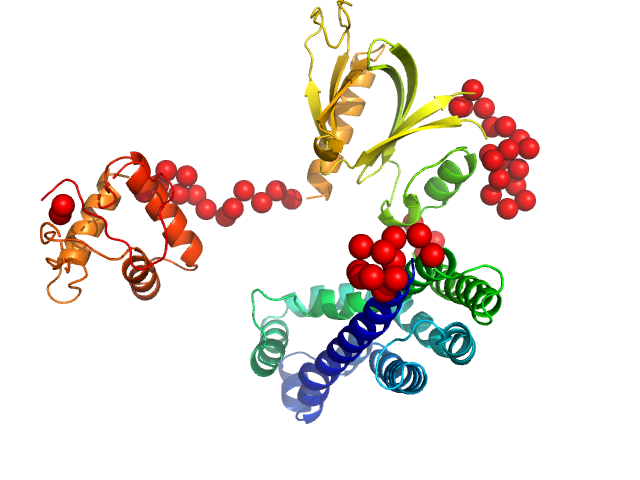
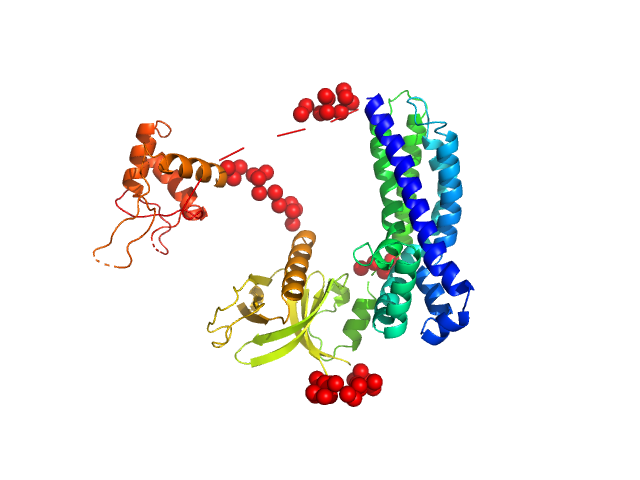
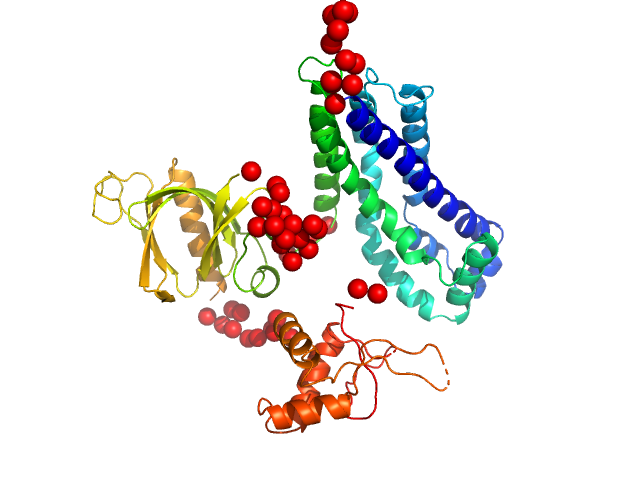
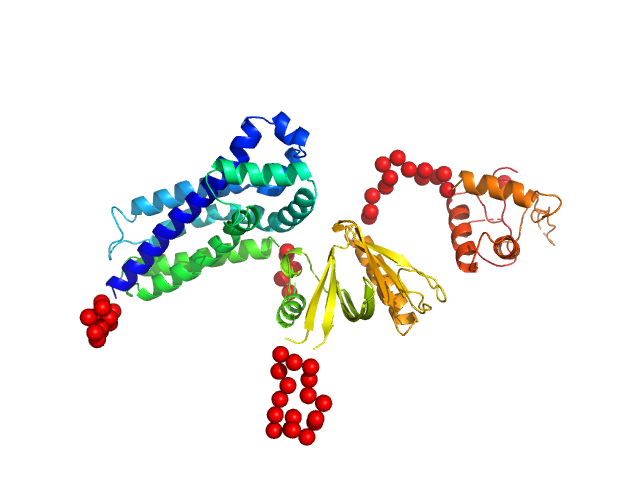
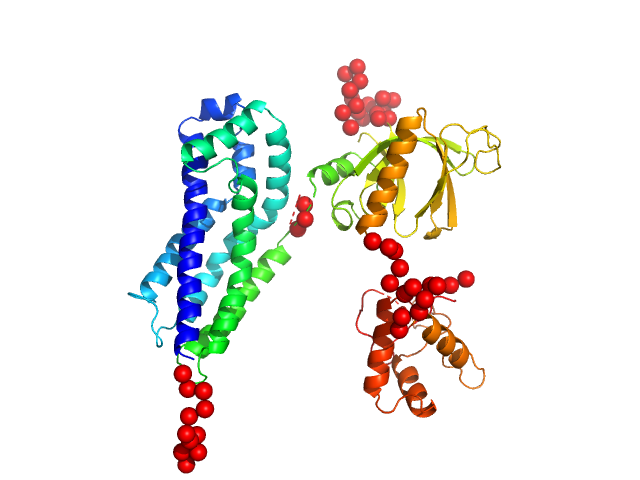
In our study, we probed into understanding how the domains in full length structure of P-Rex1 keep it inhibited by determining its structure using single particle cryo-EM and further validating it in in vitro and cell based assays. The structure showed interactions between DH and DEP1 domains and PH and IP4P domains. We incorporated mutations in these interacting interfaces. Some of the mutants in DH and DEP1 interface (L177E, I409A) that were generated in DH/PH-DEP1 fragment background were highly active. We hypothesized that these mutants would be in more open conformation and studied them using SAXS. Indeed, SAXS analysis showed that the mutants which showed highest activity sampled more elongated conformations while A170K mutant which showed decreased activity showed conformation similar to wild type protein.
|
|
 s, nm-1
s, nm-1