|
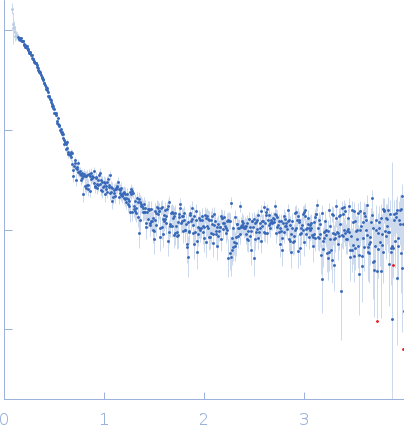
Synchrotron SAXS
data from solutions of
BAM2 in 100 mM KCl (0.5 mg/mL)
in
50 mM HEPES, 100 mM KCl, pH 7
were collected
on the
12.3.1 (SIBYLS) beam line
at the Advanced Light Source (ALS) storage ring
(Berkeley, CA, USA)
using a Pilatus3 X 2M detector
at a sample-detector distance of 2 m and
at a wavelength of λ = 0.127 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 0.50 mg/ml was measured
at 10°C.
33 successive
0.300 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Recombinant, purified BAM2 was dialyzed into 50 mM HEPES, pH 7, and 100 mM KCl at 4 ̊C overnight. Samples at a concentration of 1 mg/mL in a plate were shipped overnight on dry ice to the SIBYLS beamline at the Advanced Light Source (Dyer et al., 2014). Prior to data collection, the plate was spun at 3700 rev min-1 for 10 min. Samples were held at 10℃ during collection. The exposure was 10 s, with frames collected every 0.3 s for a total of 30 frames per sample. The detector was 1.5 m from the sample, and the beam energy was 11 keV. Matching buffer exposures were collected before and after samples to ensure there was no difference in the scattering owing to contamination of the sample cell. This setup results in scattering vectors, q, ranging from 0.013 Å-1 to 0.5 Å-1, where the scattering vector is defined as q = 4πsinθ/λ and 2θ is the measured scattering angle. Radially averaged data were processed in SCATTER (ver 4.0d) and RAW (ver. 2.1.1) to remove the scattering of the sample solvent and identify peak scattering frames (Hopkins, 2024). RAW was used to calculate the dimensions of the molecule, the molecular weight, and the pair-distance distribution function (PDDF).
|
|
 s, nm-1
s, nm-1