|
SAXS data from solutions of human DNA repair protein RAD52 homolog (amino acids 1-303) in 20 mM Bis-Tris, 10% glycerol, 400 mM NaCl, 100 mM KCl, 1 mM EDTA, pH 6 were collected were collected using a Rigaku/ BioSAXS1000 instrument at the Eppley Structural Biology Facility (University of Nebraska Medical Center, Omaha, NE, USA) equipped with a Dectris/Pilatus 100K-S detector at a sample-detector distance of 0.5 m and at a wavelength of λ = 0.154 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 7.11 mg/ml was measured at 15°C. One 9000 second frame was collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
|
|
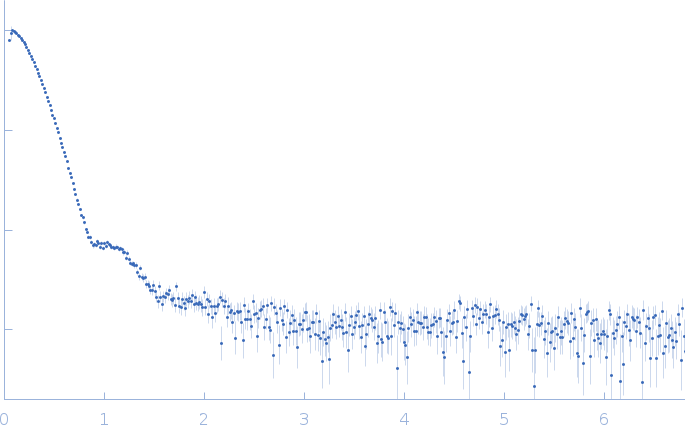 s, nm-1
s, nm-1
