|
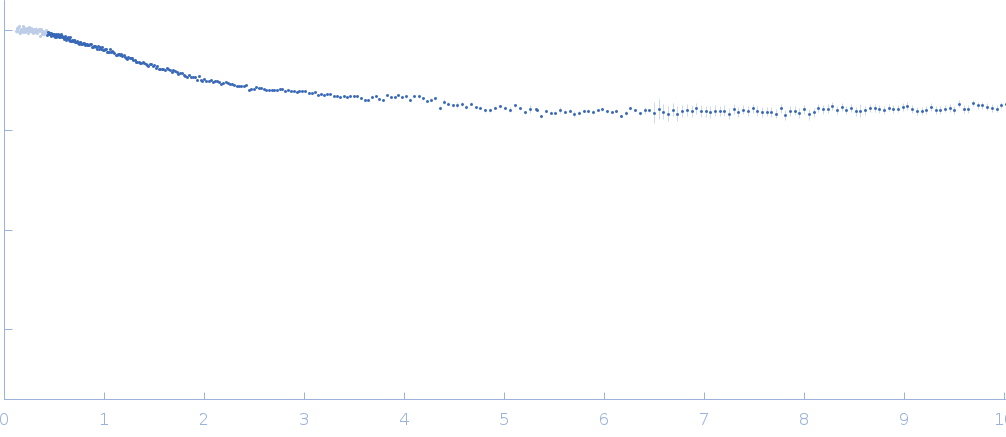
Synchrotron SAXS data from solutions of NT17 peptide in sodium phosphate buffer, pH 7.4 were collected on the TPS13A beam line at the NSRRC (Hsinchu, Taiwan) using a Eiger X 1M & 9M detector at a sample-detector distance of 2.5 m and at a wavelength of λ = 0.08265 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 0.5 and 1 mg/ml were measured at 10°C. Five successive 2 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The low angle data collected at lower concentration were merged with the highest concentration high angle data to yield the final composite scattering curve.
The sodium phosphate buffer solution contains 480 μL of 10 mM PB, pH 7.4, 16.5 μL of 100 mM NaOH, and 10 μL of 1% trifluoroacetic acid (TFA).
|
|
NT17
|
| Mol. type |
|
Protein |
| Organism |
|
synthetic construct |
| Olig. state |
|
Monomer |
| Mon. MW |
|
2.0 kDa |
| Sequence |
|
FASTA |
| |
|
 s, nm-1
s, nm-1
