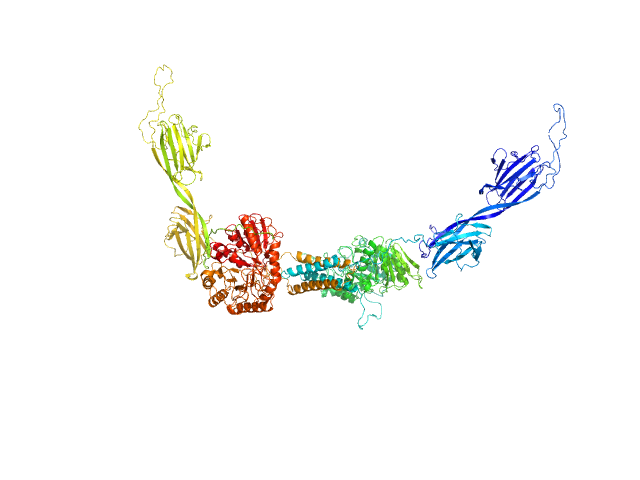
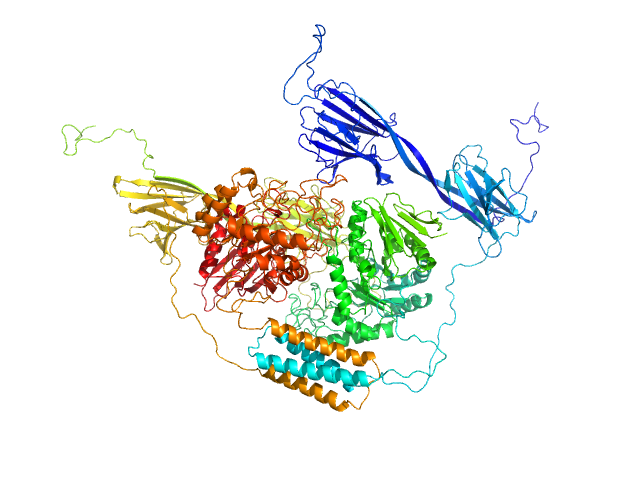
|
Synchrotron SAXS data from solutions of alpha-amylase AMY3 in 20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH 7 were collected on the 12.3.1 (SIBYLS) beam line at the Advanced Light Source (ALS; Berkeley, CA, USA) using a Pilatus3 X 2M detector at a sample-detector distance of 2.1 m and at a wavelength of λ = 0.1027 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A sample at 3 mg/ml was injected at a 0.50 ml/min flow rate onto a Shodex KW-803 column at 10°C. 20 successive 2 second frames were collected through the sample elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Cells expressing AMY3 were grown to an OD600 of approximately 0.6 at 37° C and 225 rpm, then induced with 1 mM Isopropyl-β-D-thiogalactoside (IPTG) at 20° C overnight. Cells were lysed in 50 mM Tris, pH 8.0, 100 mM sodium chloride, 0.1 mM EDTA, 1 mM Imidazole, and a Pierce complete protease inhibitor tablet using sonication at 65% power for 5 seconds on and 5 seconds off for 5 minutes. Cell debris was removed by centrifugation at 17,000 rcf at 4° C. Supernatant was then poured over 2 mL of Ni-NTA resin, and the column was allowed to drain via gravity. The column was washed with 20 mL of Wash Buffer (50 mM Tris, pH 8.0, 300 mM sodium chloride, 0.1 mM EDTA, 10 mM Imidazole). Protein was eluted with 10 mL steps from 9:1 wash buffer:elution buffer (50 mM Tris, pH 8.0, 50 mM sodium chloride, 0.1 mM EDTA, 300 mM Imidazole) to 0:10 wash buffer:elution buffer. The presence of protein was confirmed via SDS-PAGE, combined elutions were concentrated to 2 mL, and then injected onto a Sephacryl-200 16/60 column in 50 mM sodium phosphate, pH 7, 100 mM sodium chloride, and 5 mM DTT. The presence of protein in the fractions was confirmed via SDS-PAGE, protein-containing fractions were pooled, concentrated, and then stored at -80° C. Purified protein samples of AMY3 were shipped overnight at 4°C and analyzed using SEC-SAXS with in-line with UV/vis absorbance at 280nm and Multi-angle light Scattering (MALS). Radially averaged SAXS data files were processed and analyzed in Scatter IV and RAW (Hopkins, 2024).
|
|
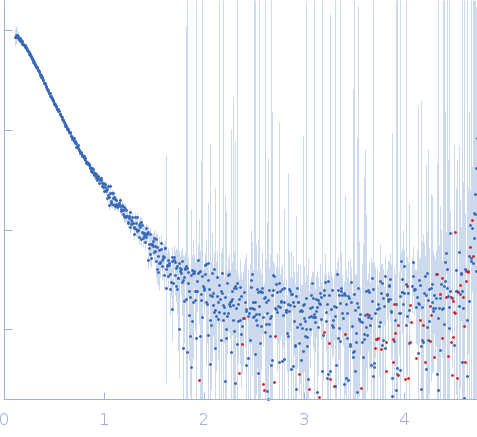 s, nm-1
s, nm-1