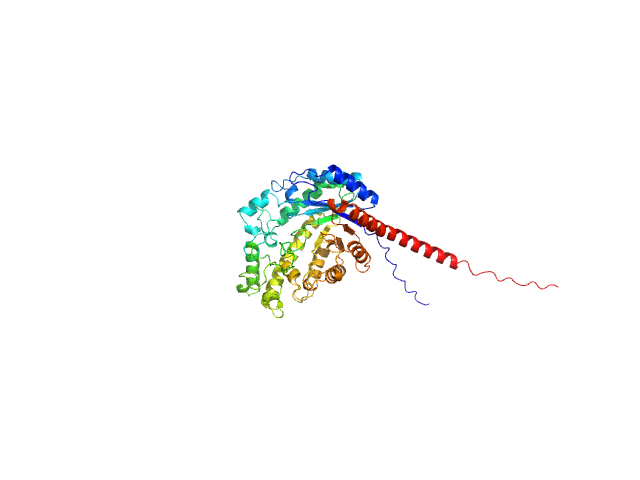
|
Synchrotron SAXS data from solutions of pseudoamylase BAM9 in 20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH 7 were collected on the 12.3.1 (SIBYLS) beam line at the Advanced Light Source (ALS; Berkeley, CA, USA) using a Pilatus3 X 2M detector at a sample-detector distance of 2.1 m and at a wavelength of λ = 0.1027 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A sample at 4 mg/ml was injected at a 0.50 ml/min flow rate onto a Shodex KW-803 column at 10°C. 20 successive 2 second frames were collected through the sample elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
BAM9 was expressed in BL21 cells with pETDuet-1-BAM9-B cultured with 100 mg/L carbenicillin. Cells were grown to an OD600 of approximately 0.6 at 37° C and 225 rpm, then induced with 1 mM Isopropyl-β-D-thiogalactoside (IPTG) at 20° C overnight. Cells were pelleted, frozen, and then sonicated on ice in 40 ml Buffer A (0.5 M NaCl, 50 mM NaH2PO4, 0.2 mM TCEP, and 2 mM imidazole, pH 8) with an EDTA-free protease-inhibitor tablet (Pierce A32965). Cell debris was removed by centrifugation at 17,000 rcf at 4° C. This supernatant was then diluted to 175 ml with Buffer A. Protein was then purified using an AKTA Start protein purification system with a 1 ml TALON cobalt-affinity column using washes of 2%, 10%, 20%, and 100% Buffer B (Cytiva Life Science, Marlborough, Massachusetts, USA). Buffer B contained 0.5 M NaCl, 50 mM NaH2PO4, 0.2 mM TCEP, and 200 mM imidazole, pH 8. Buffers A and B were supplemented with 3 mM benzamidine (TCI) when purifying AMY3 to reduce degradation. The BAM9 that was analyzed using small-angle X-ray scattering (SAXS) was additionally purified using size exclusion chromatography (SEC) with a Superdex 100 column (Cytiva). Purified protein samples of AMY3 and BAM9 alone and combined were shipped overnight at 4°C and analyzed using SEC-SAXS with in-line with UV/vis absorbance at 280nm and Multi-angle light scattering. Radially averaged SAXS data files were processed and analyzed in Scatter IV and RAW (Hopkins, 2024).
|
|
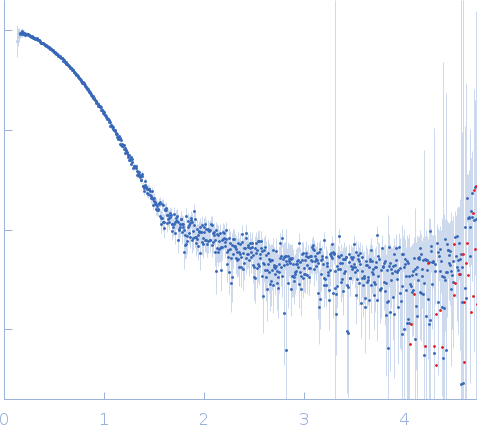 s, nm-1
s, nm-1