|
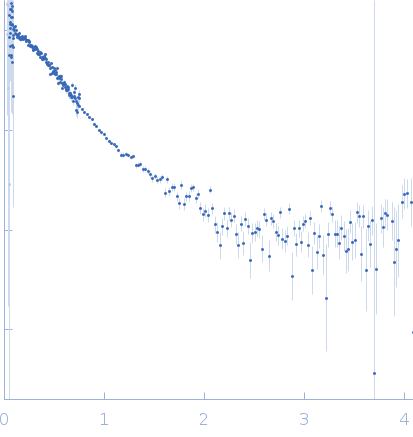
SANS data from solutions of perdeuterated N-acetylglucosamine binding Protein A (D-GbpA) in 100 mM NaCl, 20 mM Tris, 47% v/v D₂O, pH 8 were collected on the D11 beam line at the Institut Laue-Langevin (ILL; Grenoble, France). Data recorded as I(s) vs s (where s = 4πsinθ/λ, and 2θ is the scattering angle), were collected at a sample concentration of 2.4 mg/mL at 25 degrees centigrade using three instrument configurations: Set-up 1: Sample-Detector distance, 1.7 m; Collimation distance, 5.5 m; Wavelength, 0.56 nm; Frames, 1; Time per frame, 1500 s. Set-up 2: Sample-Detector distance, 8.0 m; Collimation distance, 8.0 m; Wavelength, 0.56 nm; Frames, 1; Time per frame, 1500 s. Set-up 3: Sample-Detector distance, 38.95 m; Collimation distance, 40.0 m; Wavelength, 0.56 nm; Frames, 1; Time per frame, 2100 s.
|
|
 s, nm-1
s, nm-1