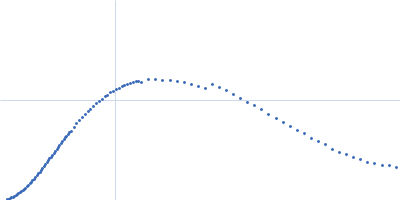
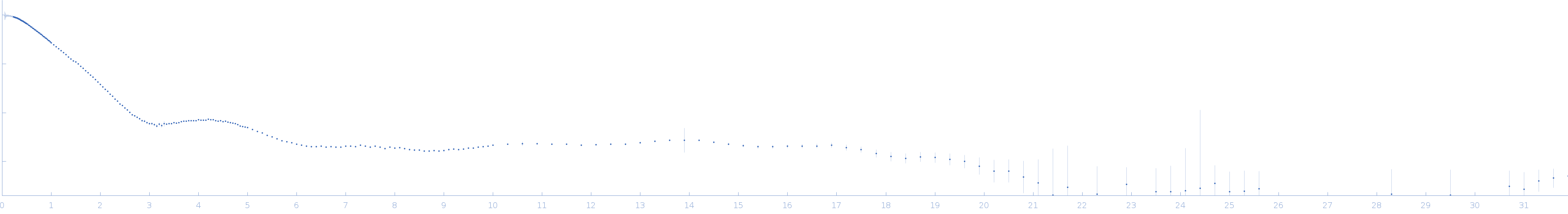
RNA triplex structures revealed by WAXS-driven MD simulations
Chen Y,
He W,
Kirmizialtin S,
Pollack L
(2022 Feb 14)
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200 mM KCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
5 mM MgCl2, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Triplex monomer, 17 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Triplex monomer, 17 kDa RNA
|
| Buffer: |
200 mM KCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Triplex monomer, 17 kDa RNA
|
| Buffer: |
5 mM MgCl2, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
10.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Long Tetraloop Hairpin Duplex monomer, 21 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Long Tetraloop Hairpin Duplex monomer, 21 kDa RNA
|
| Buffer: |
200 mM KCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Long Tetraloop Hairpin Duplex monomer, 21 kDa RNA
|
| Buffer: |
5 mM MgCl2, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Long Tetraloop Hairpin Triplex monomer, 28 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Long Tetraloop Hairpin Triplex monomer, 28 kDa RNA
|
| Buffer: |
200 mM KCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Long Tetraloop Hairpin Triplex monomer, 28 kDa RNA
|
| Buffer: |
5 mM MgCl2, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
18.2 |
nm |
|
|