Conformational flexibility of EptA driven by an interdomain helix provides insights for enzyme-substrate recognition.
Anandan A,
Dunstan NW,
Ryan TM,
Mertens HDT,
Lim KYL,
Evans GL,
Kahler CM,
Vrielink A
IUCrJ
8(Pt 5):732-746
(2021 Sep 1)
|
|
|
|
|
| Sample: |
YhbX/YhjW/YijP/YjdB family protein (L152F) monomer, 62 kDa Neisseria meningitidis serogroup … protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 0.023% n-Dodecyl β-D-maltoside (DDM), pH: 7 |
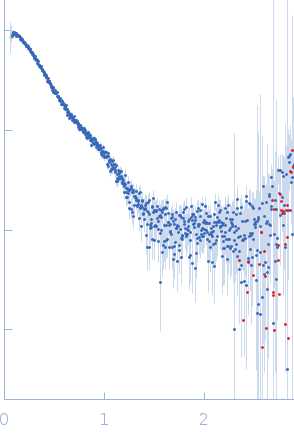
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 26
|
|
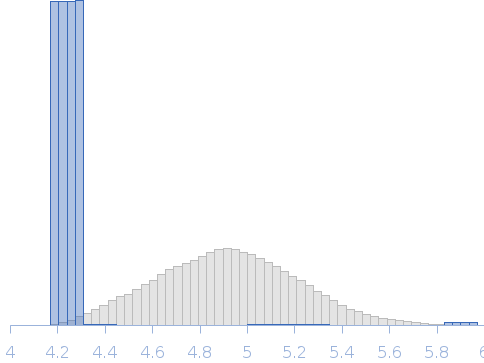
| RgGuinier |
4.2 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
304 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
YhbX/YhjW/YijP/YjdB family protein (L152F) monomer, 62 kDa Neisseria meningitidis serogroup … protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 0.14% Fos-Choline 12 (FC-12), pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 9
|
|
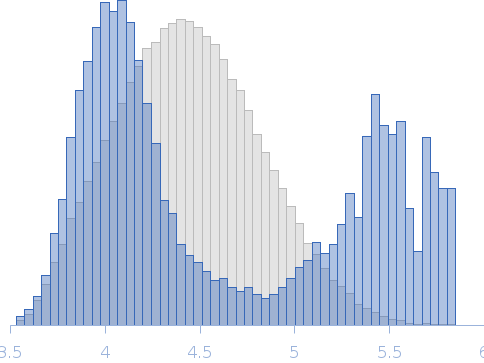
| RgGuinier |
4.6 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
186 |
nm3 |
|
|