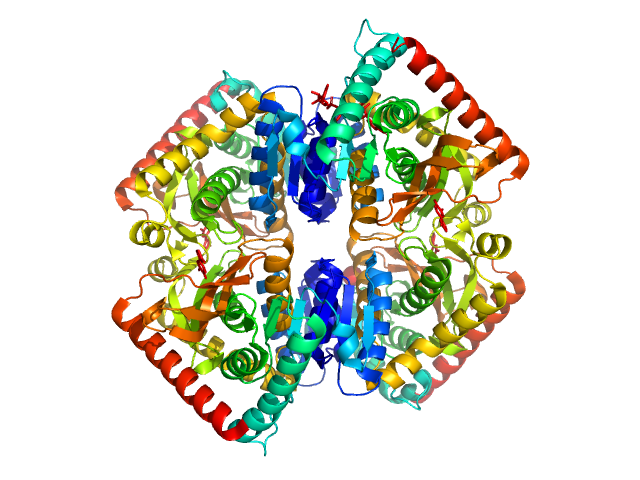
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Reyes Romero A,
Lunev S,
Popowicz G,
Calderone V,
Gentili M,
Sattler M,
Plewka J,
Taube M,
Kozak M,
Holak T,
Dömling A,
Groves M
Communications Biology
4(1)
(2021 Aug 10)
|
|
|
|
|
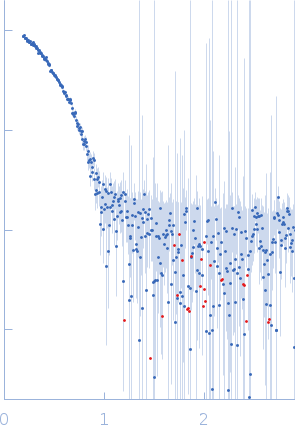
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|

|
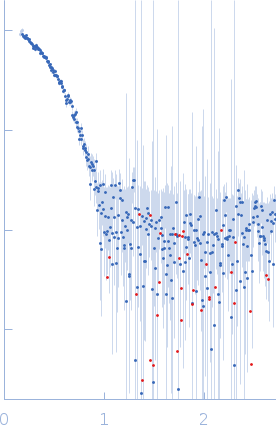
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
|
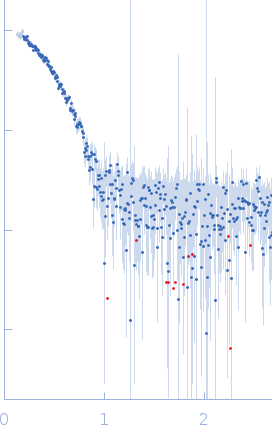
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
223 |
nm3 |
|
|