Structural basis for the inhibition of the Bacillus subtilis c-di-AMP cyclase CdaA by the phosphoglucomutase GlmM
Pathania M,
Tosi T,
Millership C,
Hoshiga F,
Morgan R,
Freemont P,
Gründling A
Journal of Biological Chemistry
:101317
(2021 Oct)
|
|
|
|
|
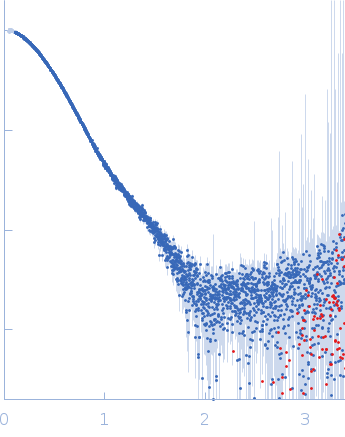
| Sample: |
Cyclic di-AMP synthase CdaA dimer, 44 kDa Bacillus subtilis (strain … protein
Phosphoglucosamine mutase dimer, 101 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 21
|
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|
|
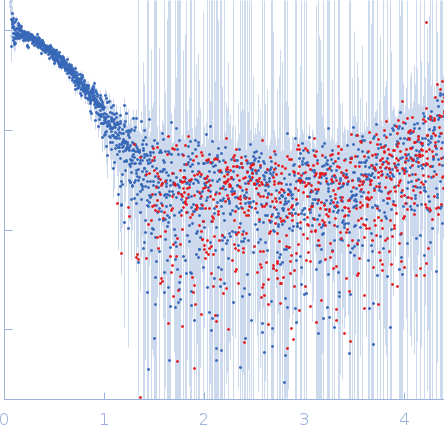
| Sample: |
Cyclic di-AMP synthase CdaA dimer, 44 kDa Bacillus subtilis (strain … protein
Phosphoglucosamine mutase dimer, 83 kDa Bacillus subtilis protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Aug 2
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoglucosamine mutase dimer, 101 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 21
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
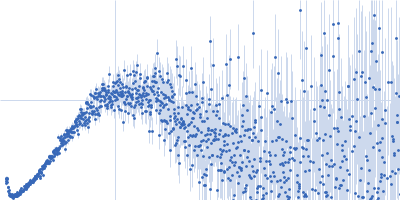
| Sample: |
Cyclic di-AMP synthase CdaA dimer, 44 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 21
|
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
60 |
nm3 |
|
|