Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Bonchuk AN,
Boyko KM,
Nikolaeva AY,
Burtseva AD,
Popov VO,
Georgiev PG
Structure
(2022 May 5)
|
|
|
|
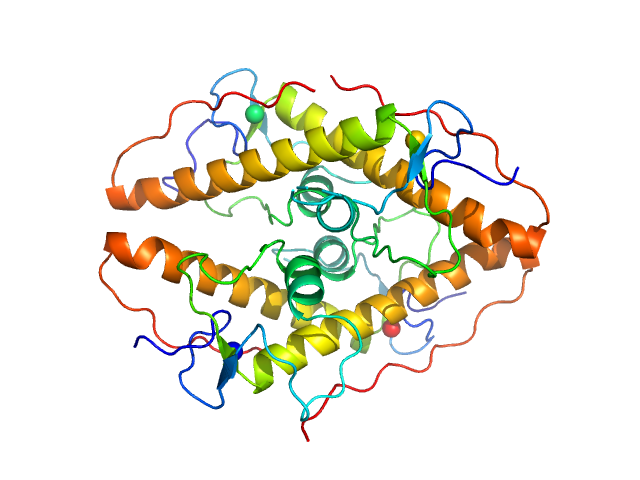
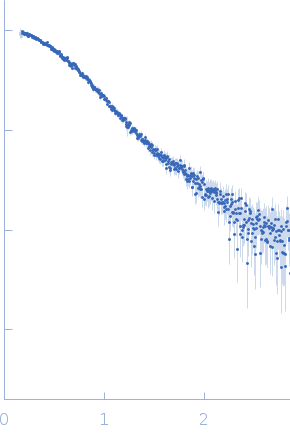
|
| Sample: |
LD15650p (Pita, isoform A) tetramer, 52 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
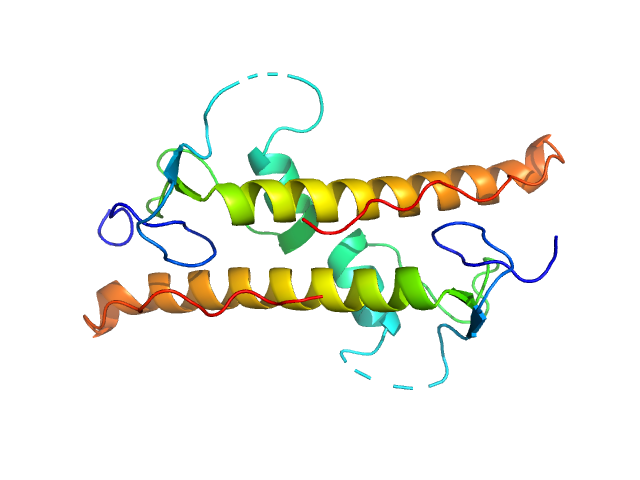
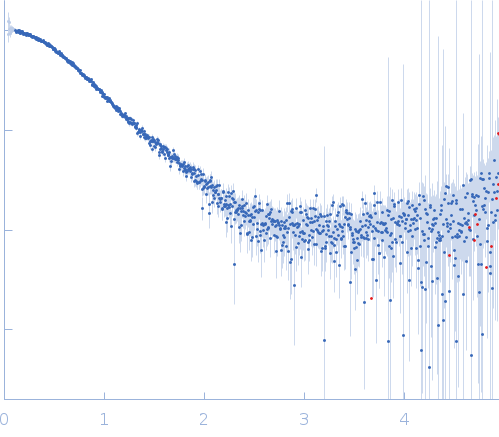
|
| Sample: |
LD15650p (Pita, isoform A; L45A) dimer, 26 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 17
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
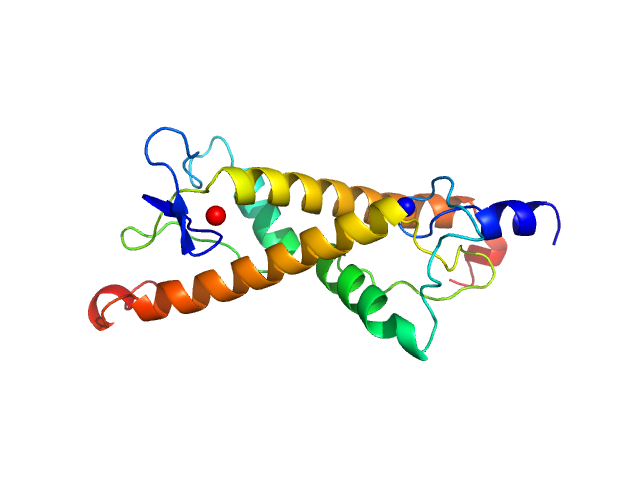
| Sample: |
LD30467p (Motif 1 binding protein) dimer, 22 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Bis-Tris-Propane, 100 mM NaCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 17
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|