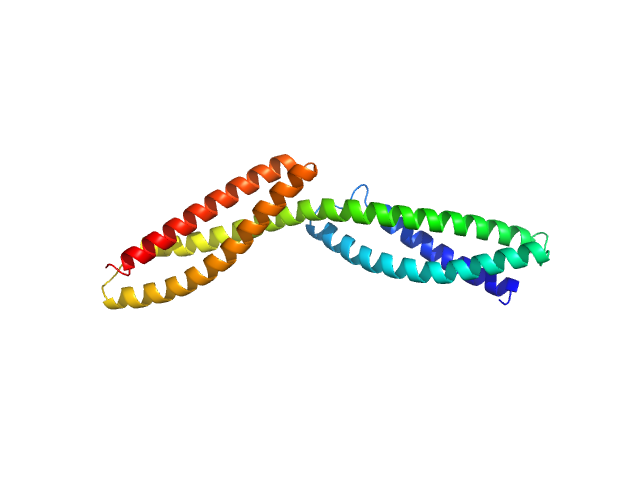
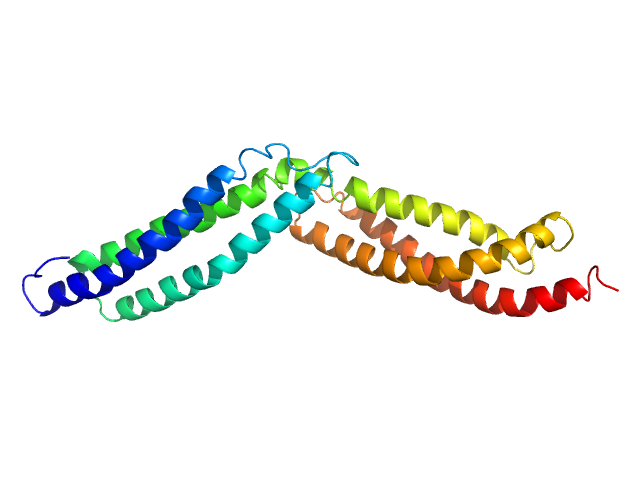
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
Delalande O,
Molza AE,
Dos Santos Morais R,
Chéron A,
Pollet É,
Raguenes-Nicol C,
Tascon C,
Giudice E,
Guilbaud M,
Nicolas A,
Bondon A,
Leturcq F,
Férey N,
Baaden M,
Perez J,
Roblin P,
Piétri-Rouxel F,
Hubert JF,
Czjzek M,
Le Rumeur E
J Biol Chem
293(18):6637-6646
(2018 May 4)
|
|
|
|
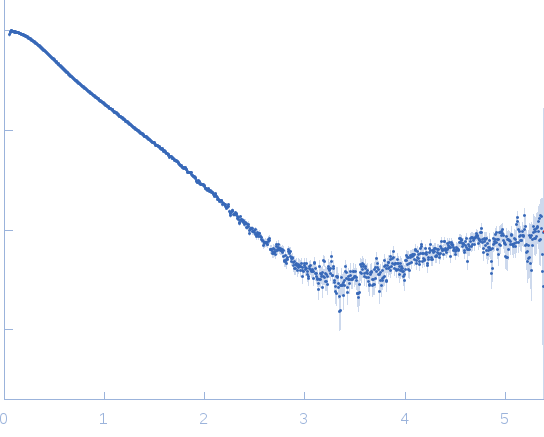
|
| Sample: |
Dystrophin central domain repeats 1 to 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Oct 7
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 1 to 3 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Sep 9
|
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 11 to 15. monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2011 Apr 11
|
|
| RgGuinier |
5.8 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 4 to 9 monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
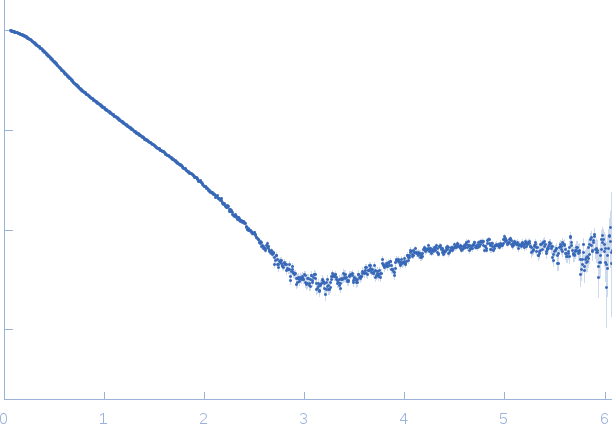
|
| Sample: |
Dystrophin central domain repeats 16 to 17. monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 16 to 19. monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 4
|
|
| RgGuinier |
4.6 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 20 to 24. monomer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Jul 10
|
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain single repeat 23 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Oct 7
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 16 to 21 (Δ2146-2305; Becker muscular dystrophy variant, deletion of exons 45-47) monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Feb 5
|
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|