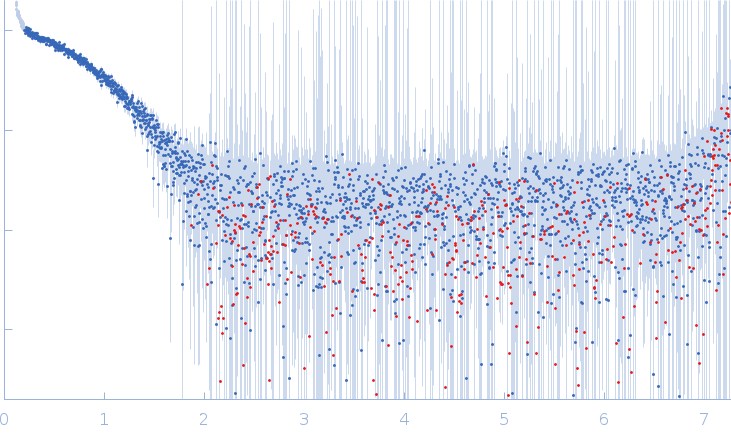
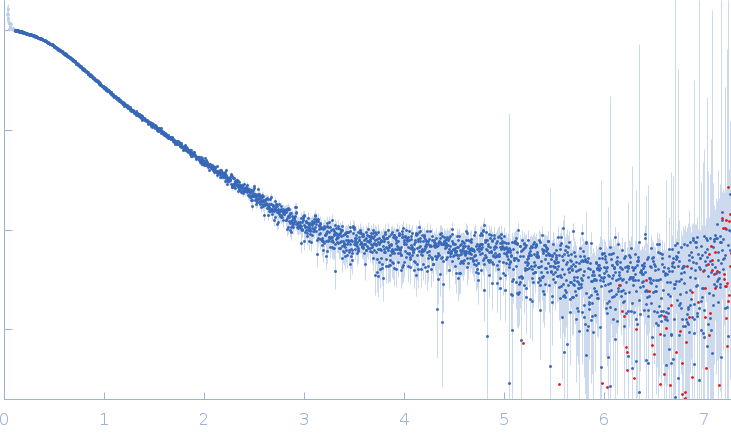
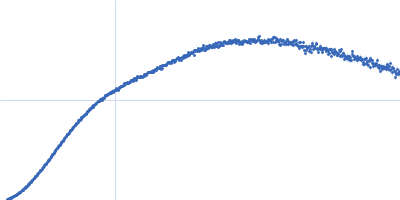
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Tants JN,
Becker LM,
McNicoll F,
Müller-McNicoll M,
Schlundt A
Nucleic Acids Res
(2022 Mar 31)
|
|
|
|
|
| Sample: |
Roquin-1 monomer, 18 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 extROQ monomer, 35 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 Nterm monomer, 51 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 Nterm Amut (K220A, K239A, R260A) monomer, 51 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 Nterm Bmut (R135E, K136E, D322A, K323A) monomer, 51 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3'UTR full-length monomer, 50 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR ADE monomer, 16 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR CDE monomer, 9 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR ADE-CDE monomer, 24 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Bulge monomer, 11 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Bulge-ADE monomer, 27 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Bulge-ADE-CDE monomer, 35 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR CDEshort monomer, 5 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
|
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Terminusuucg monomer, 10 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR ADEshort monomer, 7 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
|
| RgGuinier |
1.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|