Contactin 2 homophilic adhesion structure and conformational plasticity
Chataigner L,
Thärichen L,
Beugelink J,
Granneman J,
Mokiem N,
Snijder J,
Förster F,
Janssen B
Structure
(2023 Nov)
|
|
|
|
|
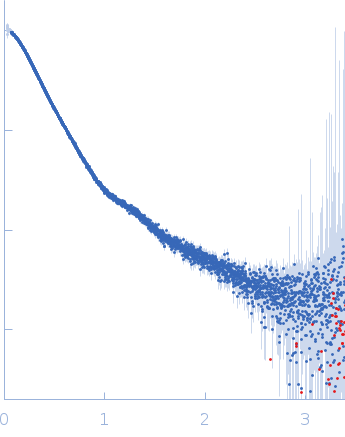
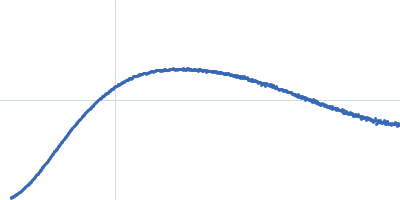
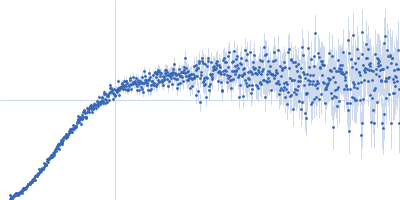
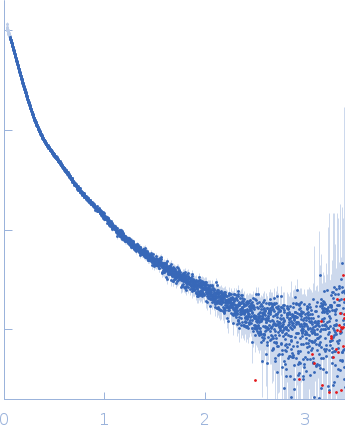
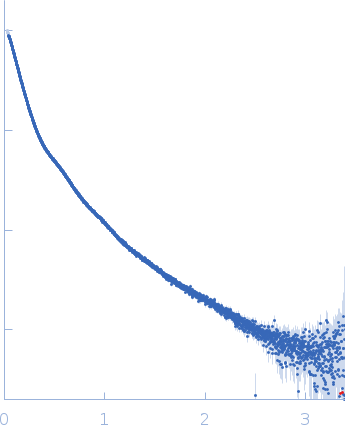
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.3 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
300 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
325 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.4 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
6.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
6.1 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
408 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.5 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
353 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.5 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
365 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
5.6 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 7
|
|
| RgGuinier |
5.8 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
412 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
9.3 |
nm |
| Dmax |
45.0 |
nm |
| VolumePorod |
780 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
10.3 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
1290 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
11.2 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
1540 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
11.0 |
nm |
| Dmax |
70.0 |
nm |
| VolumePorod |
1900 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
11.3 |
nm |
| Dmax |
66.0 |
nm |
| VolumePorod |
1820 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
12.5 |
nm |
| Dmax |
65.0 |
nm |
| VolumePorod |
2044 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
|
| RgGuinier |
14.2 |
nm |
| Dmax |
65.0 |
nm |
| VolumePorod |
2190 |
nm3 |
|
|