Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Prolič-Kalinšek M,
Volkov A,
Hadži S,
Van Dyck J,
Bervoets I,
Charlier D,
Loris R
Acta Crystallographica Section D Structural Biology
79(3):245-258
(2023 Feb 27)
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQB9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
|
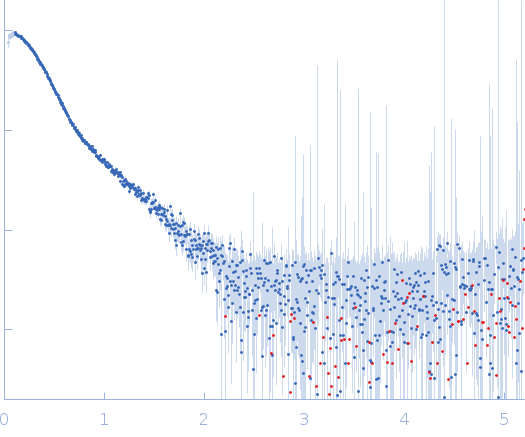
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQC9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein (mutant: L111N, F118R) monomer, 18 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Apr 14
|
|
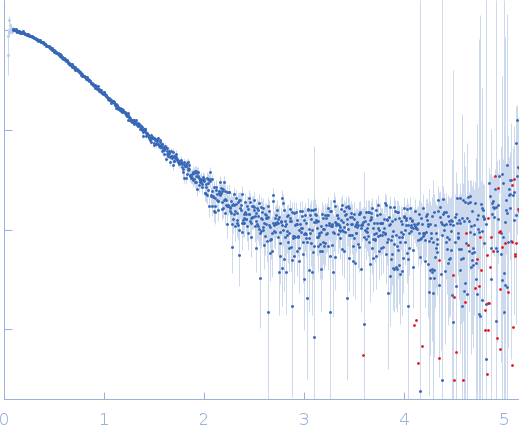
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
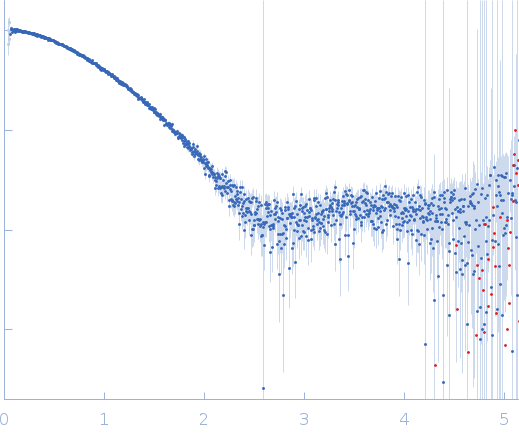
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQD9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein monomer, 13 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
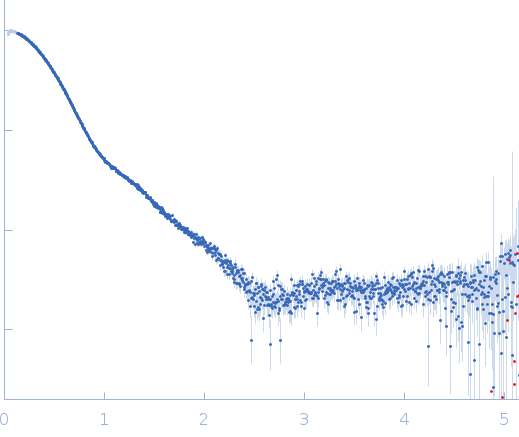
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
Om 30 base pair dsDNA dimer, 37 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 14
|
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
190 |
nm3 |
|
|

![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQB9_fit1_model1.png)
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQC9_fit1_model1.png)
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQD9_fit1_model1.png)