Atomistic structure of the SARS-CoV-2 pseudoknot in solution from SAXS-driven molecular dynamics.
He W,
San Emeterio J,
Woodside MT,
Kirmizialtin S,
Pollack L
Nucleic Acids Res
(2023 Oct 11)
|
|
|
|
|
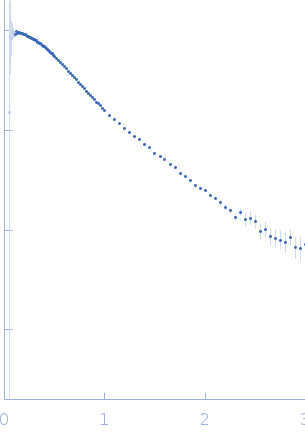
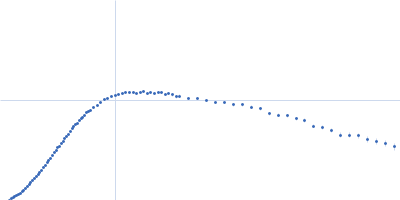
| Sample: |
Frameshifting pseudoknot from SARS CoV2, wild type monomer, 23 kDa RNA
|
| Buffer: |
50 mM MOPS, 130 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 May 15
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
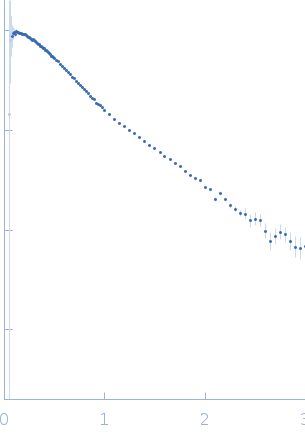
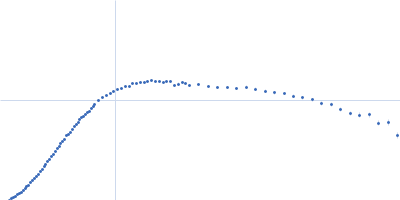
| Sample: |
Variant: non frameshifting pseudoknot from SARS CoV2 genome monomer, 23 kDa RNA
|
| Buffer: |
50 mM MOPS, 130 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 Jun 12
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
38 |
nm3 |
|
|