I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
Osipov S,
Ryzhykau Y,
Zinovev E,
Minaeva A,
Ivashchenko S,
Verteletskiy D,
Sudarev V,
Kuklina D,
Nikolaev M,
Semenov Y,
Zagryadskaya Y,
Okhrimenko I,
Gette M,
Dronova E,
Shishkin A,
Dencher N,
Kuklin A,
Ivanovich V,
Uversky V,
Vlasov A
International Journal of Molecular Sciences
24(13):10720
(2023 Jun 27)
|
|
|
|
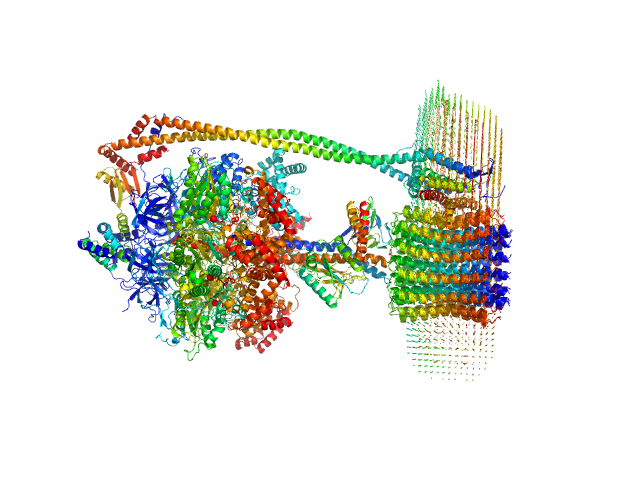
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
|
| RgGuinier |
6.6 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
927 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
150 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
|
| RgGuinier |
9.6 |
nm |
| Dmax |
41.5 |
nm |
| VolumePorod |
1506 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
250 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
|
| RgGuinier |
7.4 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
949 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRU8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
|
| RgGuinier |
7.8 |
nm |
| Dmax |
39.5 |
nm |
| VolumePorod |
1127 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRV8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
350 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
|
| RgGuinier |
8.9 |
nm |
| Dmax |
44.5 |
nm |
| VolumePorod |
1150 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRW8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
450 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
|
| RgGuinier |
10.8 |
nm |
| Dmax |
46.5 |
nm |
| VolumePorod |
1703 |
nm3 |
|
|

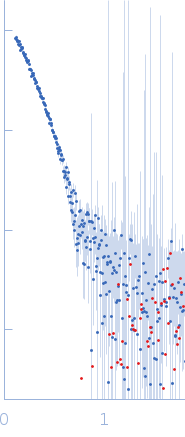

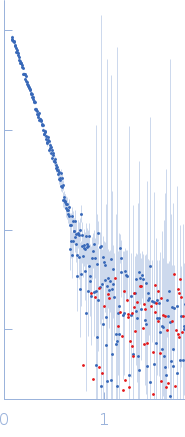
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS8_fit1_model1.png)
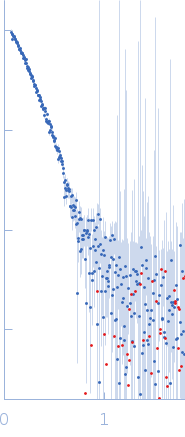
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT8_fit1_model1.png)

![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRU8_fit1_model1.png)

![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRV8_fit1_model1.png)

![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRW8_fit1_model1.png)