Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Bisello G,
Ribeiro R,
Perduca M,
Belviso B,
Polverino de' Laureto P,
Giorgetti A,
Caliandro R,
Bertoldi M
Protein Science
(2023 Jul 19)
doi: 10.1002/pro.4732
|
Submitted to SASBDB: 2023 Apr 14
Published in SASBDB:
|
|
|
|
|

|
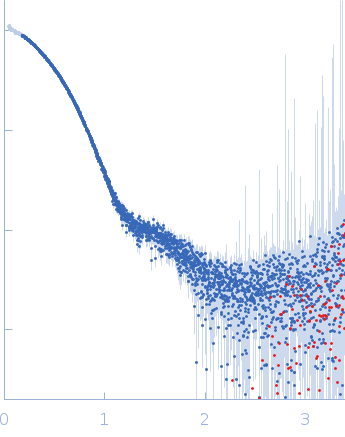
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
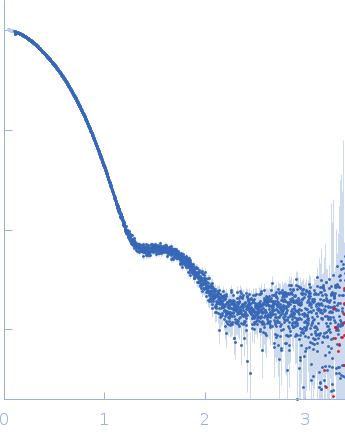
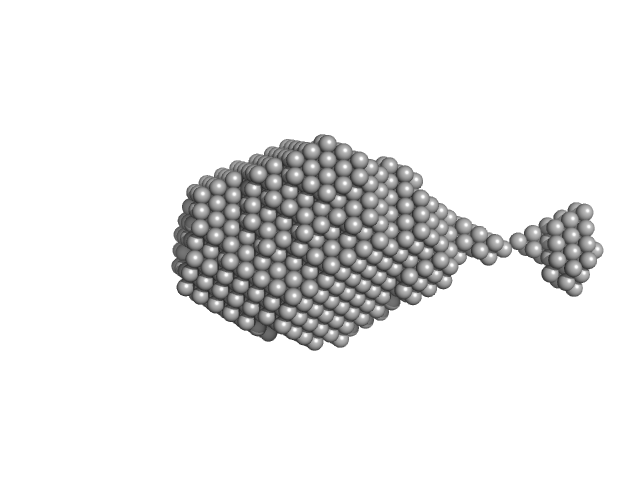
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
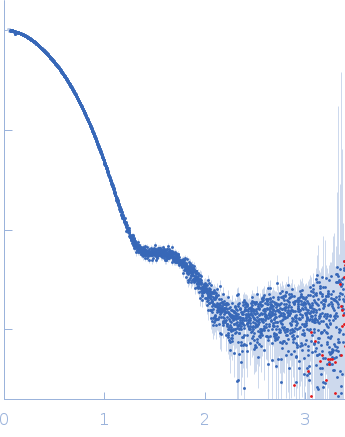
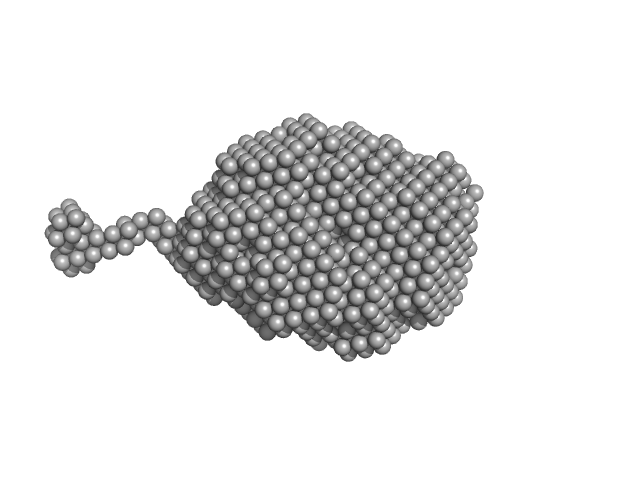
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
154 |
nm3 |
|
|