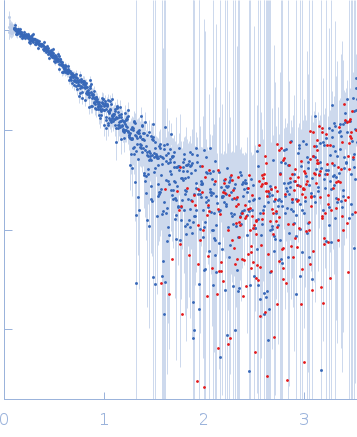
Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin
Ogbu C, Kapoor S, Vecchio AToxins 15(11):637 (2023 Oct 31)
|
doi: 10.3390/toxins15110637 |
Submitted to SASBDB: 2023 Sep 12 Published in SASBDB: |
|
|||||||||||||||||||||||
|
|||||||||||||||||||||||