The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Kocher F,
Applegate V,
Reiners J,
Port A,
Spona D,
Hänsch S,
Mirzaiebadizi A,
Ahmadian M,
Smits S,
Hegemann J,
Mölleken K
Nature Communications
15(1)
(2024 Aug 24)
|
|
|
|
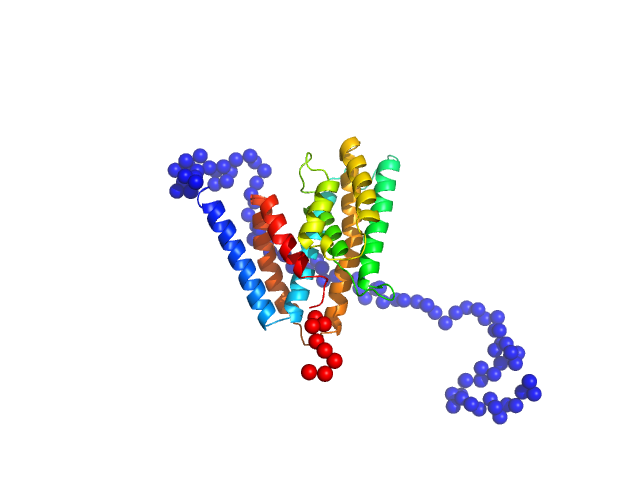
|
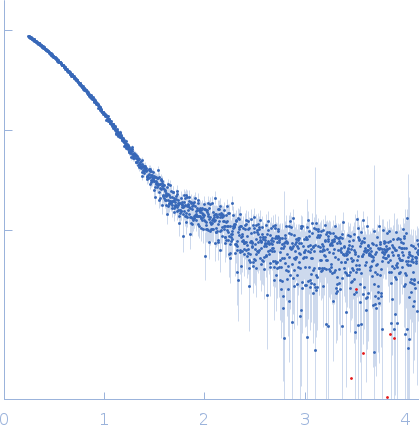
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
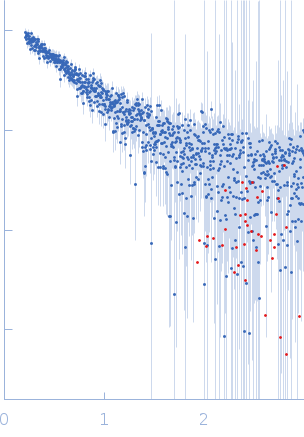
| Sample: |
Actin nucleation-promoting factor WASL monomer, 19 kDa Rattus norvegicus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
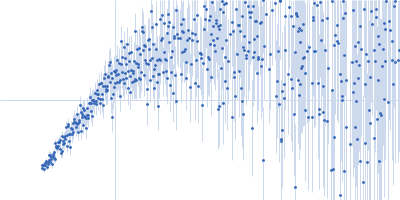
| Sample: |
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
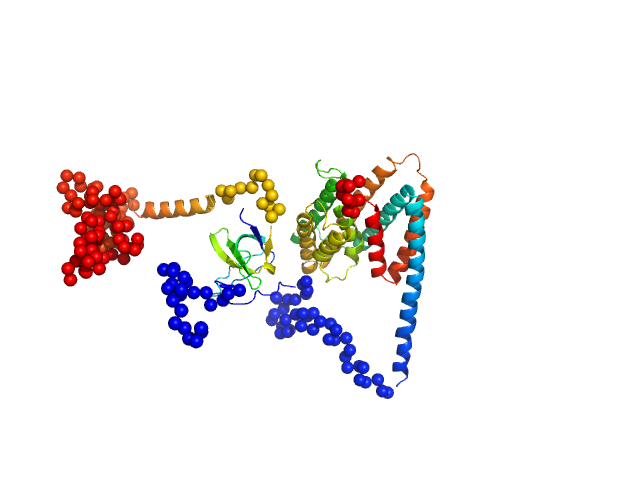
|
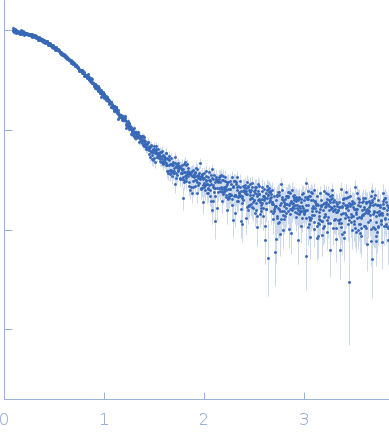
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
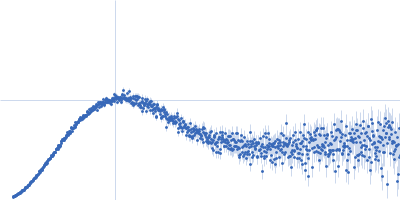
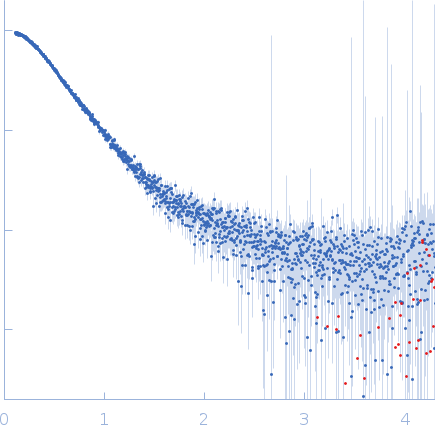
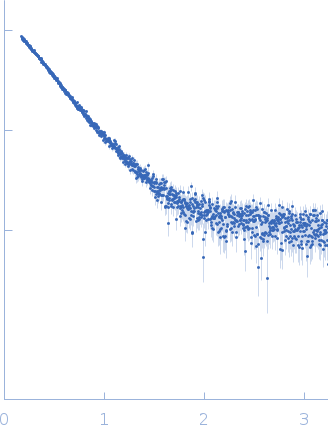
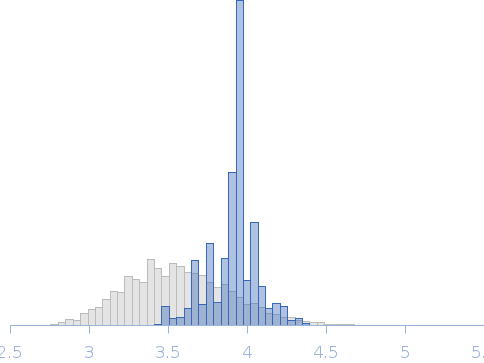
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
Actin nucleation-promoting factor WASL monomer, 19 kDa Rattus norvegicus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
102 |
nm3 |
|
|