The conformational space of RNase P RNA in solution
Lee Y,
Degenhardt M,
Skeparnias I,
Degenhardt H,
Bhandari Y,
Yu P,
Stagno J,
Fan L,
Zhang J,
Wang Y
Nature
(2024 Dec 18)
|
|
|
|
|
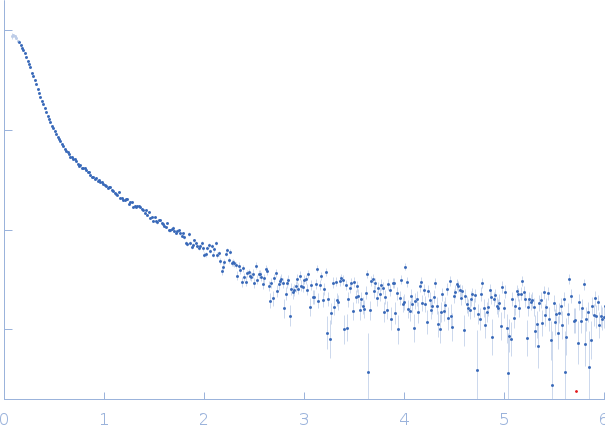
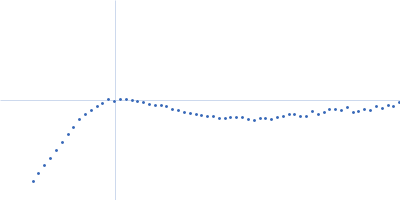
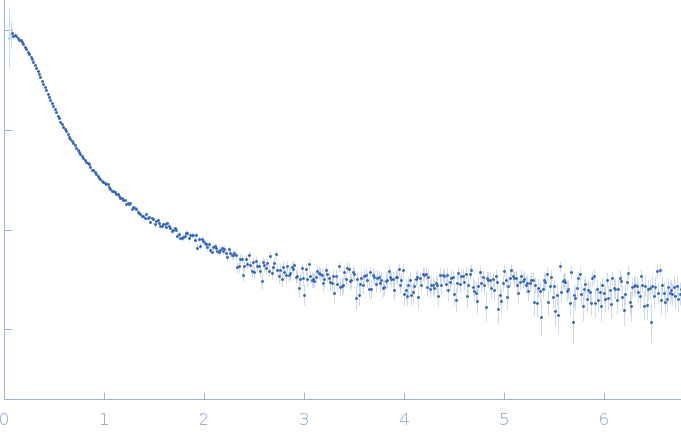
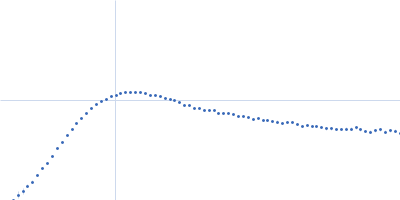
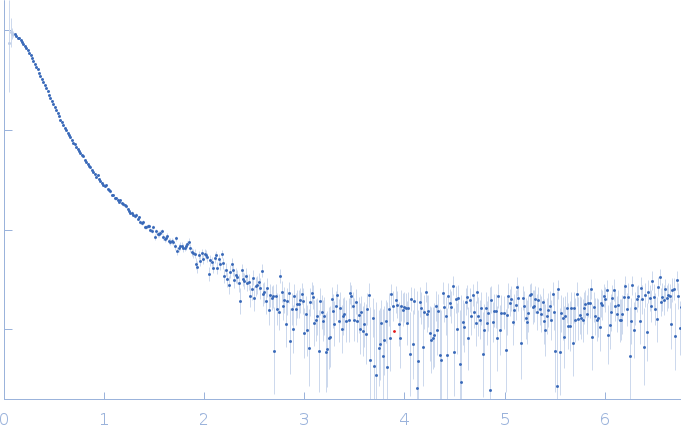
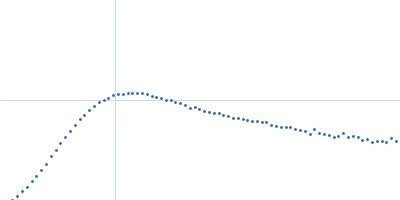
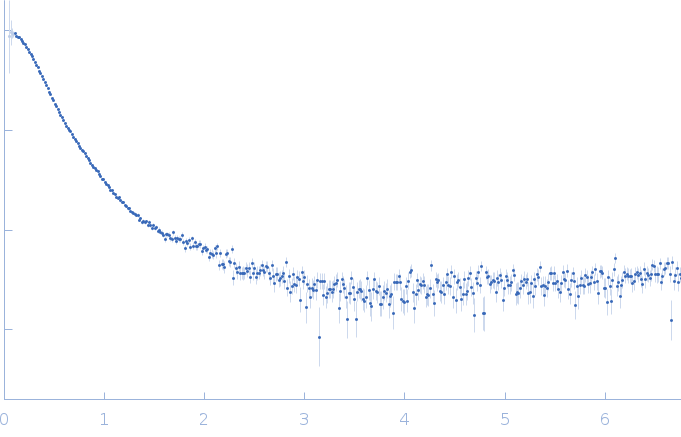
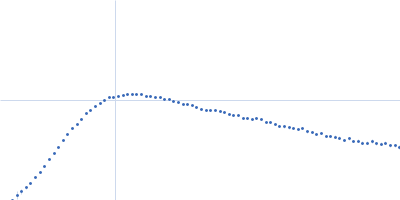
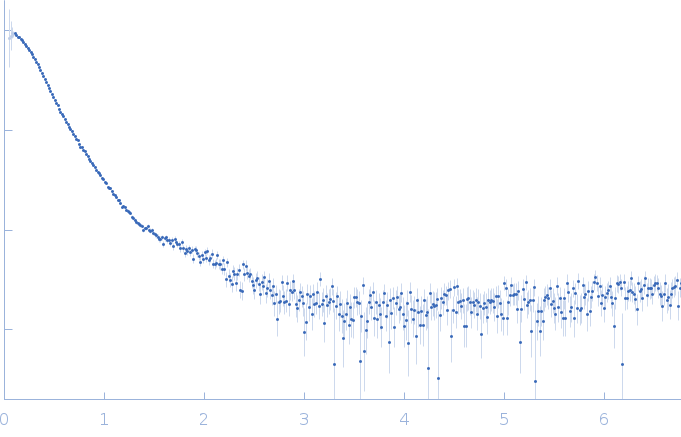
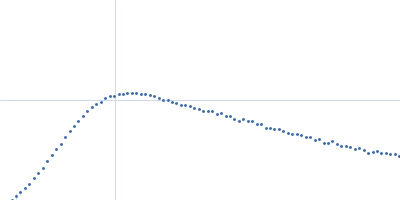
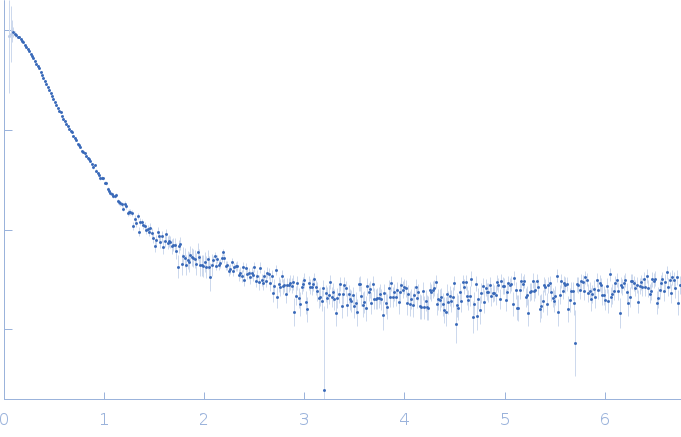
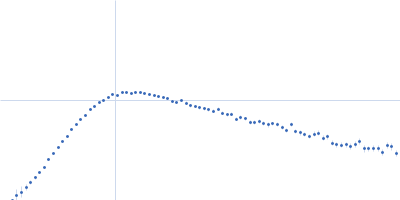
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 0.1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
813 |
nm3 |
|
|
|
|
|
|
|
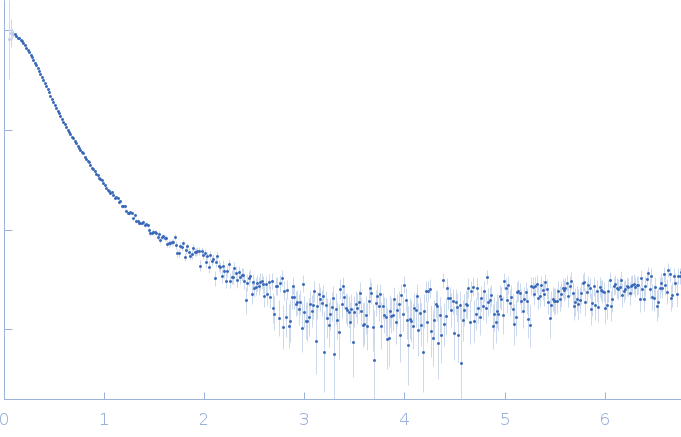
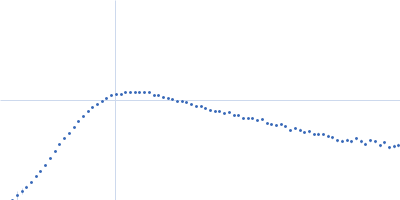
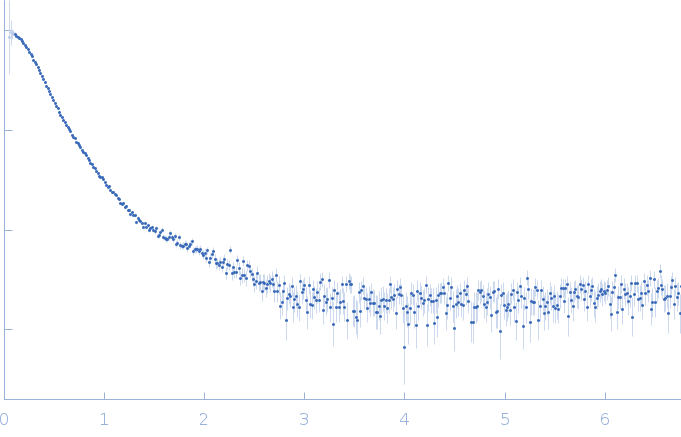
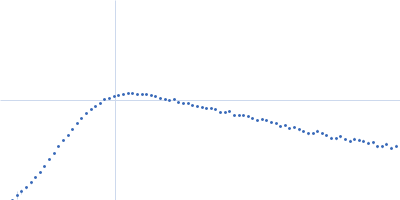
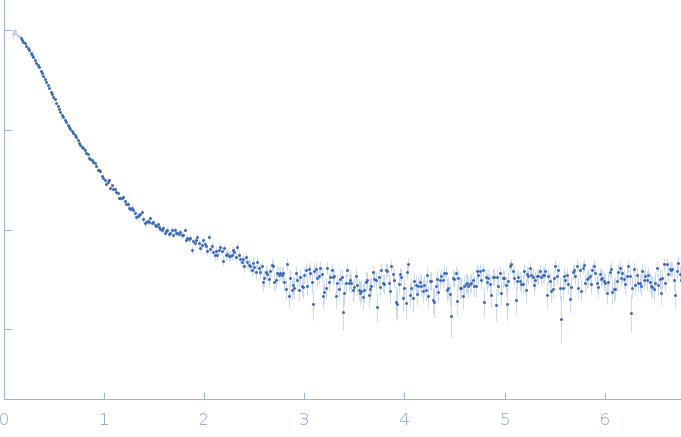
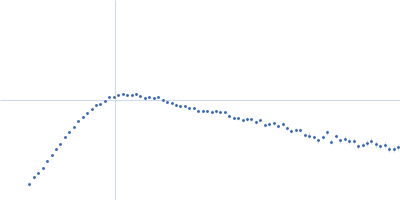
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl,1.5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
352 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 2.0 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
316 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 2.5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
306 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 3 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
301 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 8.0 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl,10.0 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
285 |
nm3 |
|
|