Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Kroupova A,
Spiteri V,
Rutter Z,
Furihata H,
Darren D,
Ramachandran S,
Chakraborti S,
Haubrich K,
Pethe J,
Gonzales D,
Wijaya A,
Rodriguez-Rios M,
Sturbaut M,
Lynch D,
Farnaby W,
Nakasone M,
Zollman D,
Ciulli A
Nature Communications
15(1)
(2024 Oct 15)
|
|
|
|
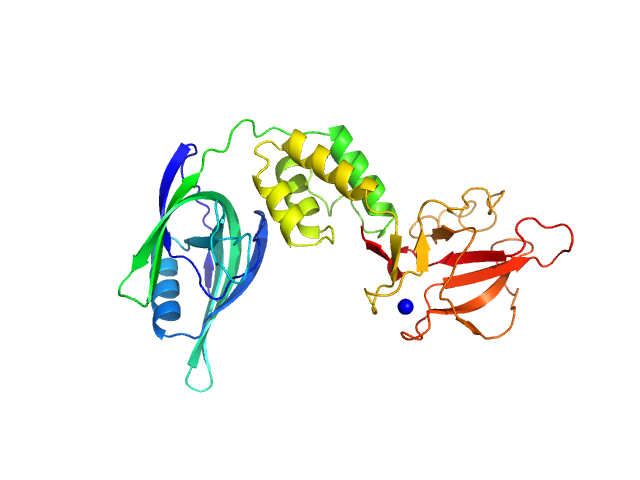
|
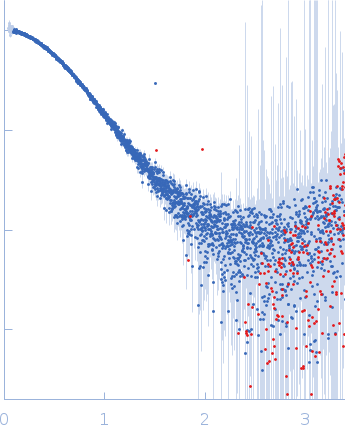
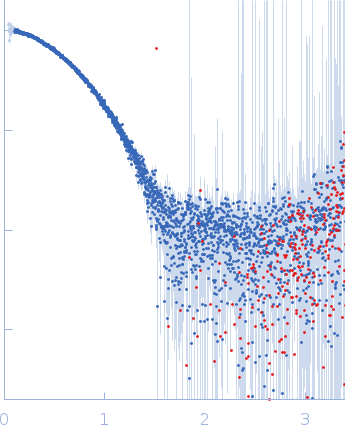
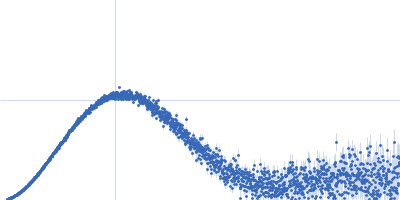
| Sample: |
Cereblon-midi monomer, 37 kDa protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
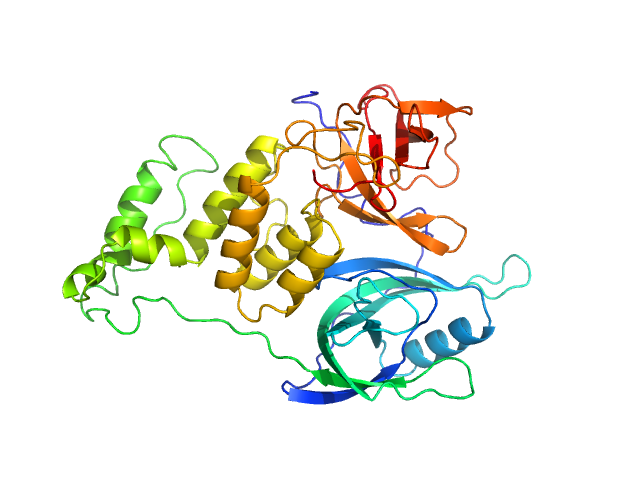
|
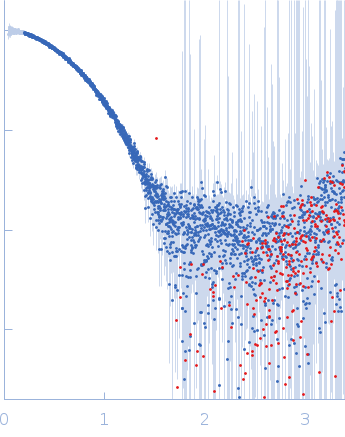
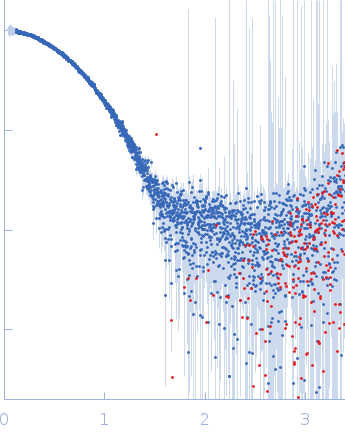
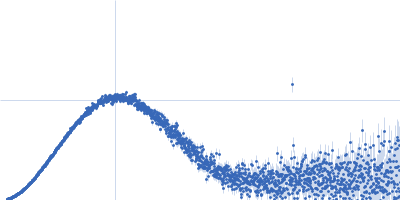
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Mezigdomide monomer, 1 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
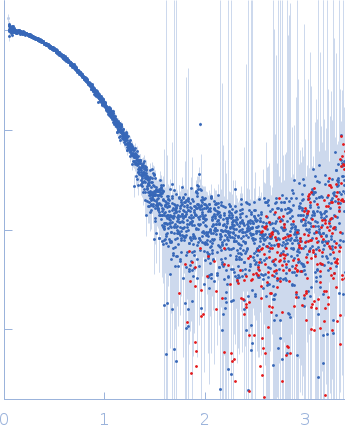
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Pomalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
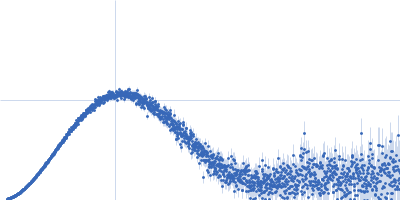
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Iberdomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Lenalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|